Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver
Abstract
:1. Introduction
2. Results and Discussion
2.1. Short Duration Exposure to the Space Environment Significantly Alters Hepatic Metabolite Profiles
2.2. Altered Betaine and Glutathione Metabolism Are Central Defects in Spaceflight Mouse Livers
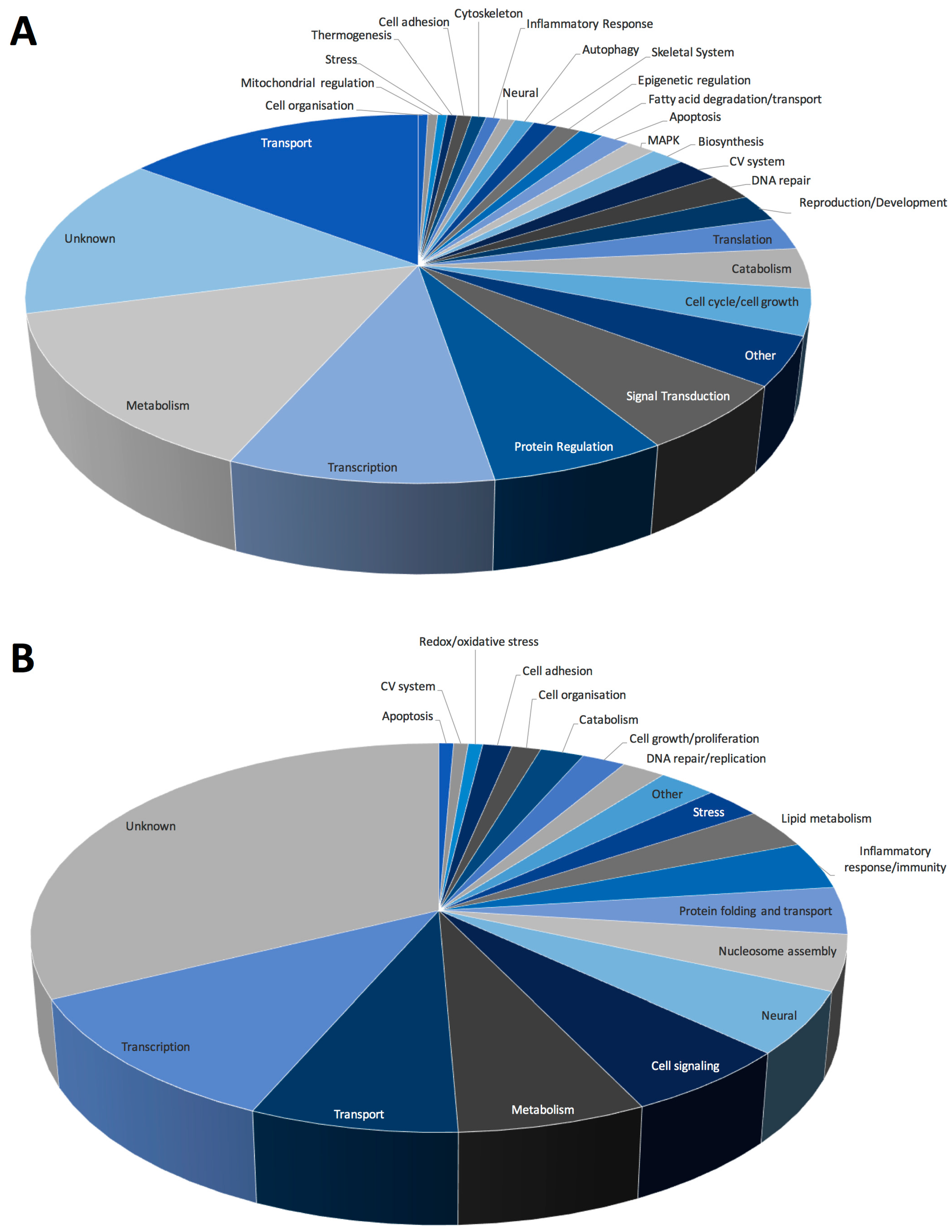
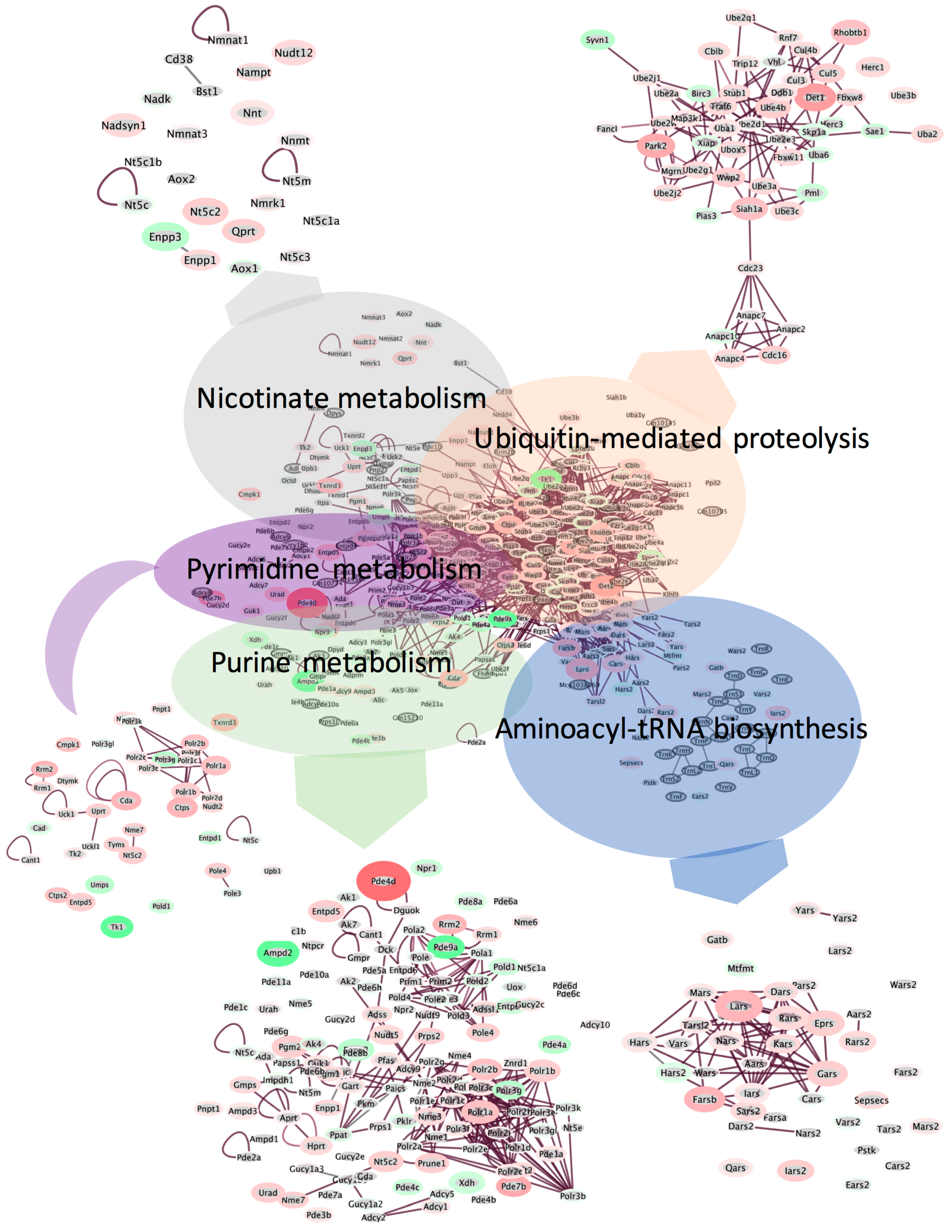
2.3. Spaceflight Causes Broad Alterations in Transcriptome Profiles in the Liver
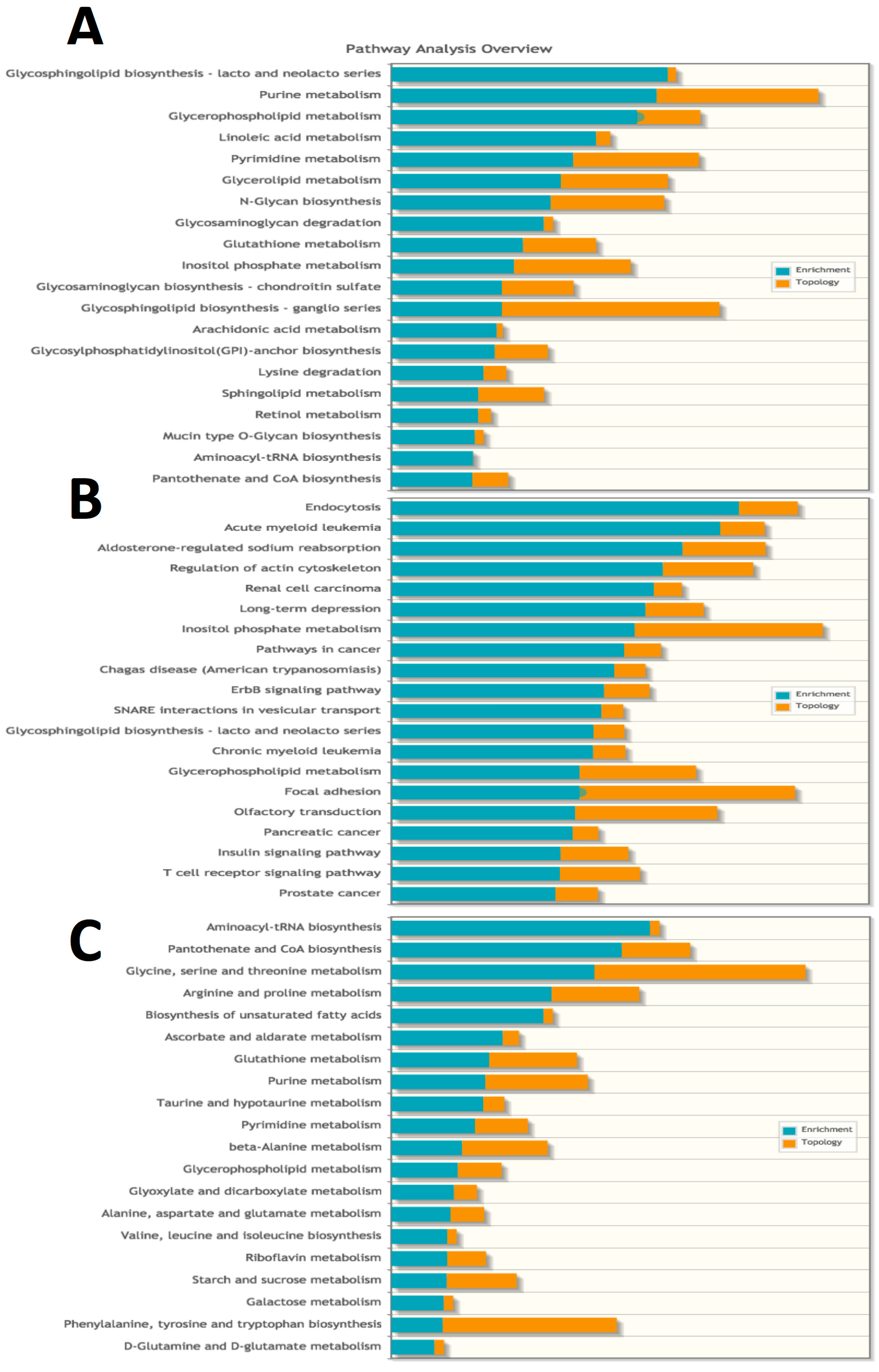
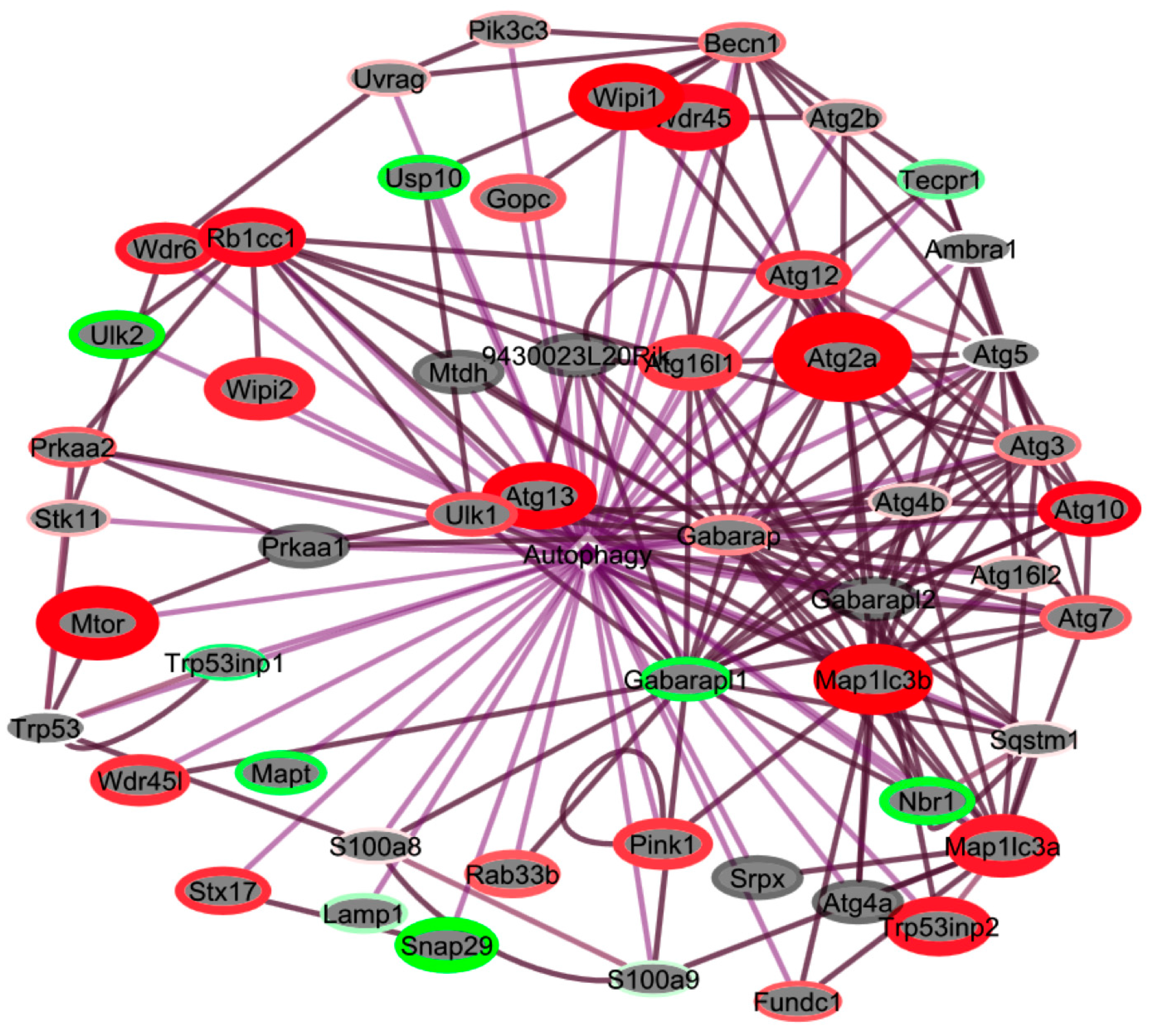
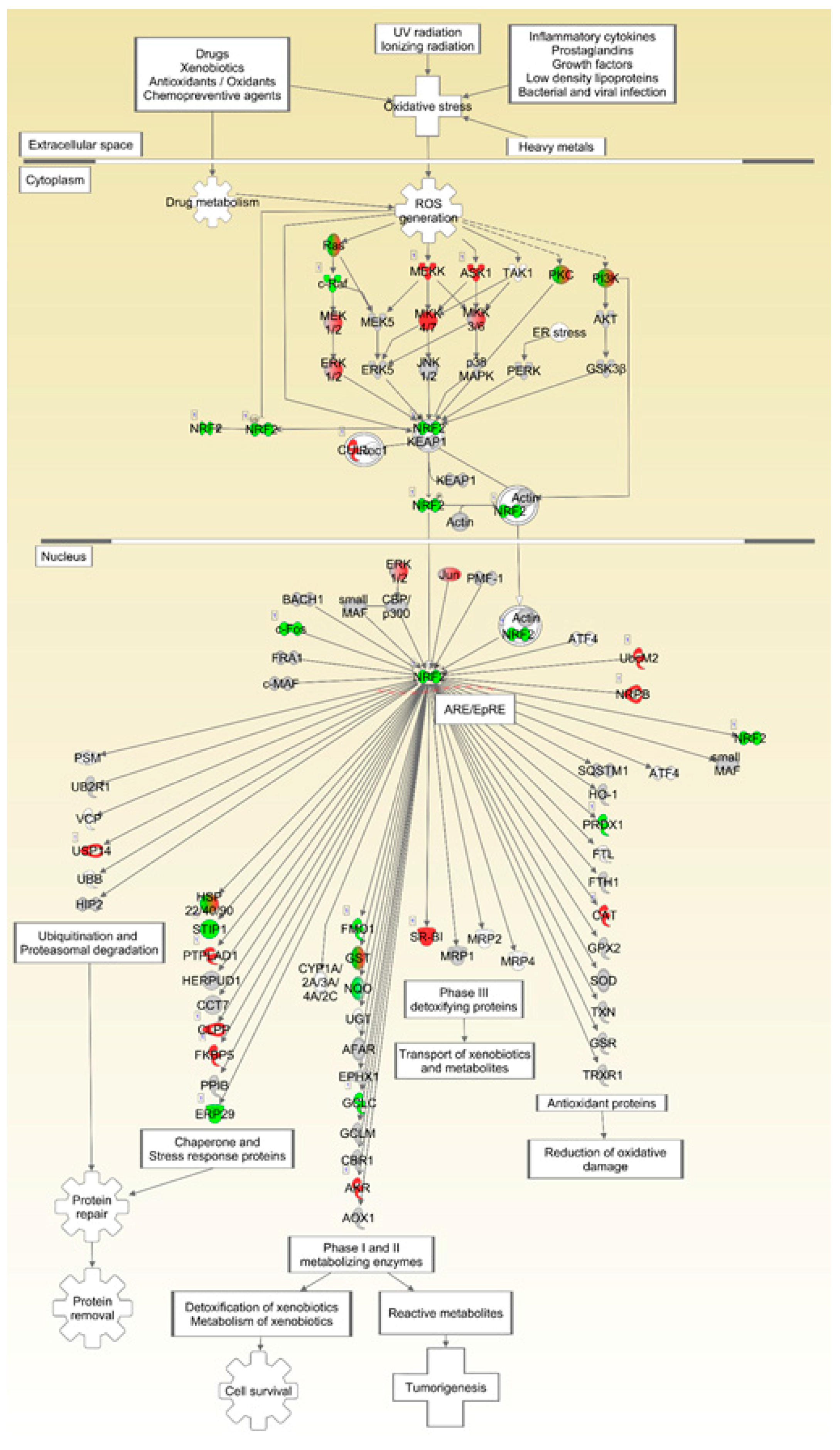
2.4. Pathways Involved in Lipid Membrane Metabolism and Protein Biosynthesis Are Enriched in Multi-‘Omics Datasets from Livers of Spaceflight Mice
3. Materials and Methods
3.1. Animals and Sample Collection
3.2. Transcriptomics
3.3. Metabolomics
3.4. Liquid Chromatography/Mass Spectrometry (LC/MS)
3.5. Gas Chromatography/Mass Spectrometry (GC/MS)
3.6. Compound Identification
3.7. Data Availability
3.8. Integrated Data Analysis
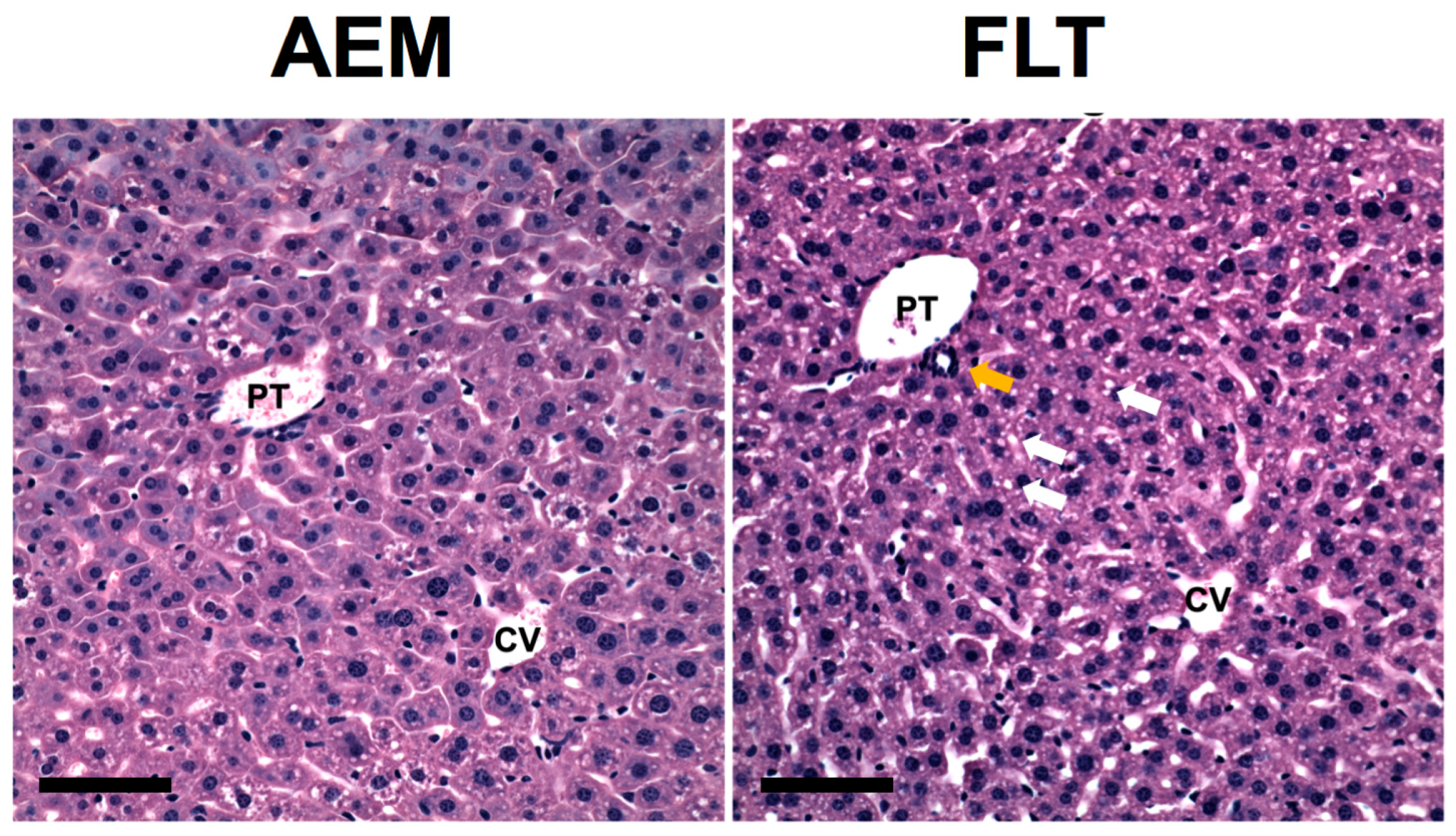
3.9. Histology
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Meehan, R.; Whitson, P.; Sams, C. The role of psychoneuroendocrine factors on spaceflight-induced immunological alterations. J. Leukoc. Biol. 1993, 54, 236–244. [Google Scholar] [PubMed]
- Alwood, J.S.; Kumar, A.; Tran, L.H.; Wang, A.; Limoli, C.L.; Globus, R.K. Low-dose, ionizing radiation and age-related changes in skeletal microarchitecture. J. Aging Res. 2011, 2012. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; Schimmerling, W.; Wilson, J.W.; Peterson, L.E.; Badhwar, G.D.; Saganti, P.B.; Dicello, J.F. Space radiation cancer risks and uncertainties for mars missions. Radiat. Res. 2001, 156, 682–688. [Google Scholar] [CrossRef]
- Pecaut, M.J.; Mao, X.W.; Bellinger, D.L.; Jonscher, K.R.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Mohney, R.P.; Gridley, D.S. Is spaceflight-induced immune dysfunction linked to systemic changes in metabolism? PLoS ONE 2017, 12, e0174174. [Google Scholar] [CrossRef] [PubMed]
- Jonscher, K.R.; Alfonso-Garcia, A.; Suhalim, J.L.; Orlicky, D.J.; Potma, E.O.; Ferguson, V.L.; Bouxsein, M.L.; Bateman, T.A.; Stodieck, L.S.; Levi, M.; et al. Spaceflight activates lipotoxic pathways in mouse liver. PLoS ONE 2016, 11, e0152877. [Google Scholar]
- Gobbel, G.T.; Bellinzona, M.; Vogt, A.R.; Gupta, N.; Fike, J.R.; Chan, P.H. Response of postmitotic neurons to X-irradiation: Implications for the role of DNA damage in neuronal apoptosis. J. Neurosci. 1998, 18, 147–155. [Google Scholar] [PubMed]
- Shinohara, C.; Gobbel, G.T.; Lamborn, K.R.; Tada, E.; Fike, J.R. Apoptosis in the subependyma of young adult rats after single and fractionated doses of x-rays. Cancer Res. 1997, 57, 2694–2702. [Google Scholar] [PubMed]
- Bellinzona, M.; Gobbel, G.T.; Shinohara, C.; Fike, J.R. Apoptosis is induced in the subependyma of young adult rats by ionizing irradiation. Neurosci. Lett. 1996, 208, 163–166. [Google Scholar] [CrossRef]
- Jafari, M.; Salehi, M.; Zardooz, H.; Rostamkhani, F. Response of liver antioxidant defense system to acute and chronic physical and psychological stresses in male rats. EXCLI J. 2014, 13, 161–171. [Google Scholar] [PubMed]
- Duda, W.; Curzytek, K.; Kubera, M.; Iciek, M.; Kowalczyk-Pachel, D.; Bilska-Wilkosz, A.; Lorenc-Koci, E.; Leskiewicz, M.; Basta-Kaim, A.; Budziszewska, B.; et al. The effect of chronic mild stress and imipramine on the markers of oxidative stress and antioxidant system in rat liver. Neurotox. Res. 2016, 30, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.W.; Pecaut, M.J.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Bouxsein, M.L.; Gridley, D.S. Biological and metabolic response in STS-135 space-flown mouse skin. Free Radic. Res. 2014, 48, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.W.; Pecaut, M.J.; Stodieck, L.S.; Ferguson, V.L.; Bateman, T.A.; Bouxsein, M.; Jones, T.A.; Moldovan, M.; Cunningham, C.E.; Chieu, J.; et al. Spaceflight environment induces mitochondrial oxidative damage in ocular tissue. Radiat. Res. 2013, 180, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Baqai, F.P.; Gridley, D.S.; Slater, J.M.; Luo-Owen, X.; Stodieck, L.S.; Ferguson, V.; Chapes, S.K.; Pecaut, M.J. Effects of spaceflight on innate immune function and antioxidant gene expression. J. Appl. Physiol. 2009, 106, 1935–1942. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.Y.; Wang, Y.; Vanhoutte, P.M. Senescence of cultured porcine coronary arterial endothelial cells is associated with accelerated oxidative stress and activation of NFKB. J. Vasc. Res. 2010, 47, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Limoli, C.; Searby, N.D.; Almeida, E.A.; Loftus, D.J.; Vercoutere, W.; Morey-Holton, E.; Giedzinski, E.; Mojarrab, R.; Hilton, D.; et al. Shared oxidative pathways in response to gravity-dependent loading and γ-irradiation of bone marrow-derived skeletal cell progenitors. Radiats. Biol. Radioecol. 2007, 47, 281–285. [Google Scholar] [PubMed]
- Evans, J.L.; Goldfine, I.D.; Maddux, B.A.; Grodsky, G.M. Oxidative stress and stress-activated signaling pathways: A unifying hypothesis of type 2 diabetes. Endocr. Rev. 2002, 23, 599–622. [Google Scholar] [CrossRef] [PubMed]
- Madamanchi, N.R.; Vendrov, A.; Runge, M.S. Oxidative stress and vascular disease. Arterioscler Arterioscler. Thromb. Vasc. Biol. 2005, 25, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Csanyi, G.; Miller, F.J., Jr. Oxidative stress in cardiovascular disease. Int. J. Mol. Sci. 2014, 15, 6002–6008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva, M.S.; Zimmerman, P.M.; Meguid, M.M.; Nandi, J.; Ohinata, K.; Xu, Y.; Chen, C.; Tada, T.; Inui, A. Anorexia in space and possible etiologies: An overview. Nutrition 2002, 18, 805–813. [Google Scholar] [CrossRef]
- Tobin, B.W.; Uchakin, P.N.; Leeper-Woodford, S.K. Insulin secretion and sensitivity in space flight: Diabetogenic effects. Nutrition 2002, 18, 842–848. [Google Scholar] [CrossRef]
- Dello, S.A.; Neis, E.P.; de Jong, M.C.; van Eijk, H.M.; Kicken, C.H.; Olde, D.S.W.; Dejong, C.H. Systematic review of ophthalmate as a novel biomarker of hepatic glutathione depletion. Clin. Nutr. 2013, 32, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Anselm, V.; Novikova, S.; Zgoda, V. Re-adaption on earth after spaceflights affects the mouse liver proteome. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Chandler, T.L.; White, H.M. Choline and methionine differentially alter methyl carbon metabolism in bovine neonatal hepatocytes. PLoS ONE 2017, 12, e0171080. [Google Scholar] [CrossRef] [PubMed]
- Lobley, G.E.; Connell, A.; Revell, D. The importance of transmethylation reactions to methionine metabolism in sheep: Effects of supplementation with creatine and choline. Br. J. Nutr. 1996, 75, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Levillain, O.; Marescau, B.; Possemiers, I.; de Deyn, P. Dehydration modifies guanidino compound concentrations in the different zones of the rat kidney. Pflugers Arch. 2002, 444, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Levillain, O.; Marescau, B.; Possemiers, I.; al Banchaabouchi, M.; de Deyn, P.P. Influence of 72% injury in one kidney on several organs involved in guanidino compound metabolism: A time course study. Pflugers Arch. 2001, 442, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Nordgren, M.; Fransen, M. Peroxisomal metabolism and oxidative stress. Biochimie 2014, 98, 56–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fransen, M.; Nordgren, M.; Wang, B.; Apanasets, O. Role of peroxisomes in ROS/RNS-metabolism: Implications for human disease. Biochim. Biophys. Acta 2012, 1822, 1363–1373. [Google Scholar] [CrossRef] [PubMed]
- Bonekamp, N.A.; Volkl, A.; Fahimi, H.D.; Schrader, M. Reactive oxygen species and peroxisomes: Struggling for balance. Biofactors 2009, 35, 346–355. [Google Scholar] [CrossRef] [PubMed]
- Antonenkov, V.D.; Grunau, S.; Ohlmeier, S.; Hiltunen, J.K. Peroxisomes are oxidative organelles. Antioxid. Redox Signal. 2010, 13, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Manivannan, S.; Scheckhuber, C.Q.; Veenhuis, M.; van der Klei, I.J. The impact of peroxisomes on cellular aging and death. Front. Oncol. 2012, 2, 50. [Google Scholar] [CrossRef] [PubMed]
- Zayzafoon, M.; Meyers, V.E.; McDonald, J.M. Microgravity: The immune response and bone. Immunol. Rev. 2005, 208, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.T.; Keen, C.L.; Gershwin, M.E. Microgravity and immune responsiveness: Implications for space travel. Nutrition 2002, 18, 889–898. [Google Scholar] [CrossRef]
- Pecaut, M.J.; Nelson, G.A.; Peters, L.L.; Kostenuik, P.J.; Bateman, T.A.; Morony, S.; Stodieck, L.S.; Lacey, D.L.; Simske, S.J.; Gridley, D.S. Genetic models in applied physiology: Selected contribution: Effects of spaceflight on immunity in the C57BL/6 mouse. I. Immune population distributions. J. Appl. Physiol. 2003, 94, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.A.; Wiesmann, W.; Kidwell, W.; Cannon, T.; Kerns, L.; Serke, C.; Delaplaine, T.; Pranger, A.; Lee, K.P. Effect of spaceflight on human stem cell hematopoiesis: Suppression of erythropoiesis and myelopoiesis. J. Leukoc. Biol. 1996, 60, 69–76. [Google Scholar] [PubMed]
- Bikman, B.T.; Summers, S.A. Ceramides as modulators of cellular and whole-body metabolism. J. Clin. Investig. 2011, 121, 4222–4230. [Google Scholar] [CrossRef] [PubMed]
- Kasumov, T.; Li, L.; Li, M.; Gulshan, K.; Kirwan, J.P.; Liu, X.; Previs, S.; Willard, B.; Smith, J.D.; McCullough, A. Ceramide as a mediator of non-alcoholic fatty liver disease and associated atherosclerosis. PLoS ONE 2015, 10, e0126910. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Sloan, K.E. The mitochondrial epitranscriptome: The roles of RNA modifications in mitochondrial translation and human disease. Cell Mol. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vare, V.Y.; Eruysal, E.R.; Narendran, A.; Sarachan, K.L.; Agris, P.F. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Dewe, J.M.; Fuller, B.L.; Lentini, J.M.; Kellner, S.M.; Fu, D. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol. Cell Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gonzalez, C.; Moukadiri, I.; Villarroya, M.; Lopez-Pascual, E.; Tuck, S.; Armengod, M.E. Mutations in the caenorhabditis elegans orthologs of human genes required for mitochondrial tRNA modification cause similar electron transport chain defects but different nuclear responses. PLoS Genet 2017, 13, e1006921. [Google Scholar] [CrossRef] [PubMed]
- Paquette, J.; Tokuyasu, T. Egan: Exploratory gene association networks. Bioinformatics 2010, 26, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Akizu, N.; Cantagrel, V.; Schroth, J.; Cai, N.; Vaux, K.; McCloskey, D.; Naviaux, R.K.; van Vleet, J.; Fenstermaker, A.G.; Silhavy, J.L.; et al. AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 2013, 154, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Hudoyo, A.W.; Hirase, T.; Tandelillin, A.; Honda, M.; Shirai, M.; Cheng, J.; Morisaki, H.; Morisaki, T. Role of AMPD2 in impaired glucose tolerance induced by high fructose diet. Mol. Genet. Metab. Rep. 2017, 13, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Monticone, M.; Liu, Y.; Pujic, N.; Cancedda, R. Activation of nervous system development genes in bone marrow derived mesenchymal stem cells following spaceflight exposure. J. Cell. Biochem. 2010, 111, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Dvorochkin, N.; Torres, M.L.; Yousuf, R.; Burns, B.P.; Globus, R.K.; Almeida, E.A. Mechanical unloading of bone in microgravity reduces mesenchymal and hematopoietic stem cell-mediated tissue regeneration. Stem Cell Res. 2014, 13, 181–201. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Dvorochkin, N.; Lee, C.; Alwood, J.S.; Yousuf, R.; Pianetta, P.; Globus, R.K.; Burns, B.P.; Almeida, E.A. Microgravity induces pelvic bone loss through osteoclastic activity, osteocytic osteolysis, and osteoblastic cell cycle inhibition by CDKN1A/P21. PLoS ONE 2013, 8, e61372. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.; Sato, K.; Almeida, E.A. Stem cell health and tissue regeneration in microgravity. Stem Cells Dev. 2014, 23, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Blaber, E.A.; Finkelstein, H.; Dvorochkin, N.; Sato, K.Y.; Yousuf, R.; Burns, B.P.; Globus, R.K.; Almeida, E.A. Microgravity reduces the differentiation and regenerative potential of embryonic stem cells. Stem Cells Dev. 2015, 24, 2605–2621. [Google Scholar] [CrossRef] [PubMed]
- Chanfreau, G.F. Impact of RNA modifications and RNA-modifying enzymes on eukaryotic ribonucleases. Enzymes 2017, 41, 299–329. [Google Scholar] [PubMed]
- Kraft, C.; Peter, M.; Hofmann, K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 2010, 12, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Smedley, D.; Haider, S.; Durinck, S.; Pandini, L.; Provero, P.; Allen, J.; Arnaiz, O.; Awedh, M.H.; Baldock, R.; Barbiera, G.; et al. The biomart community portal: An innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015, 43, W589–W598. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Yepes, J.; Burns, M.; Anandhan, A.; Khalimonchuk, O.; del Razo, L.M.; Quintanilla-Vega, B.; Pappa, A.; Panayiotidis, M.I.; Franco, R. Oxidative stress, redox signaling, and autophagy: Cell death versus survival. Antioxid. Redox Signal. 2014, 21, 66–85. [Google Scholar] [CrossRef] [PubMed]
- Kriegenburg, F.; Poulsen, E.G.; Koch, A.; Kruger, E.; Hartmann-Petersen, R. Redox control of the ubiquitin-proteasome system: From molecular mechanisms to functional significance. Antioxid. Redox Signal. 2011, 15, 2265–2299. [Google Scholar] [CrossRef] [PubMed]
- Wang, K. Autophagy and apoptosis in liver injury. Cell Cycle 2015, 14, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Liver regeneration. J. Cell Physiol. 2007, 213, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Advances in liver regeneration. Expert. Rev. Gastroenterol. Hepatol. 2014, 8, 897–907. [Google Scholar] [CrossRef] [PubMed]
- Toshima, T.; Shirabe, K.; Fukuhara, T.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Okano, S.; Maehara, Y. Suppression of autophagy during liver regeneration impairs energy charge and hepatocyte senescence in mice. Hepatology 2014, 60, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Diehl, A.M.; Chute, J. Underlying potential: Cellular and molecular determinants of adult liver repair. J. Clin. Investig. 2013, 123, 1858–1860. [Google Scholar] [CrossRef] [PubMed]
- Ogrodnik, M.; Miwa, S.; Tchkonia, T.; Tiniakos, D.; Wilson, C.L.; Lahat, A.; Day, C.P.; Burt, A.; Palmer, A.; Anstee, Q.M.; et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun. 2017, 8, 15691. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulou, M.; Gordillo, R.; Koliaki, C.; Gancheva, S.; Jelenik, T.; Herder, C.; Markgraf, D.; Scherer, P.E.; Roden, M. Serum and hepatic sphingolipids relate to insulin resistance, hepatic mitochondrial capacity and oxidative stress in non-alcoholic fatty liver disease. In Diabetologie und Stoffwechsel; Thieme: New York, NY, USA, 2017; pp. S1–S84. [Google Scholar]
- Meikle, P.J.; Summers, S.A. Sphingolipids and phospholipids in insulin resistance and related metabolic disorders. Nat. Rev. Endocrinol. 2017, 13, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Dehaven, C.D.; Evans, A.M.; Dai, H.; Lawton, K.A. Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J. Cheminform. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55. [Google Scholar] [CrossRef]
- Russell, T.D.; Palmer, C.A.; Orlicky, D.J.; Fischer, A.; Rudolph, M.C.; Neville, M.C.; McManaman, J.L. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: Roles of adipophilin and lipid metabolism. J. Lipid. Res. 2007, 48, 1463–1475. [Google Scholar] [CrossRef] [PubMed]



| Biochemicals | FC | log2 (FC) | p | −log10 (p) |
|---|---|---|---|---|
| Reduced | ||||
| 4-Guanidinobutanoate 1,* | 0.377 | −1.409 | 1.98 × 10−5 | 4.703 |
| Glycerophosphorylcholine * | 0.282 | −1.826 | 1.52 × 10−3 | 2.817 |
| 3-Ureidopropionate | 0.466 | −1.102 | 3.83 × 10−2 | 1.417 |
| Increased | ||||
| 3-hydroxybutyrate * | 3.229 | 1.691 | 7.54 × 10−5 | 4.123 |
| Glutarate pentanedioate * | 4.766 | 2.253 | 8.40 × 10−5 | 4.076 |
| Propionylcarnitine * | 2.440 | 1.287 | 1.68 × 10−4 | 3.776 |
| 3-methylglutarylcarnitine * | 3.478 | 1.798 | 5.90 × 10−4 | 3.229 |
| Dimethylglycine * | 2.195 | 1.134 | 1.87 × 10−3 | 2.728 |
| Hexadecanedioate * | 2.189 | 1.131 | 2.07 × 10−3 | 2.684 |
| Ophthalmate | 2.326 | 1.218 | 1.30 × 10−2 | 1.887 |
| Hydroxyisovaleroyl carnitine | 2.567 | 1.360 | 1.37 × 10−2 | 1.862 |
| Putrescine | 2.857 | 1.514 | 3.48 × 10−2 | 1.459 |
| Cholate | 2.602 | 1.380 | 4.98 × 10−2 | 1.302 |
| Taurodeoxycholate | 2.978 | 1.574 | 5.00 × 10−2 | 1.301 |
| Intake | AEM a | FLT | FLT/AEM | p-Value |
|---|---|---|---|---|
| Food Intake (g) b | 4.08 ± 0.10 | 4.09 ± 0.18 | 1.00 | 0.865 |
| Water Intake (g) | 3.38 ± 0.22 | 2.73 ± 0.01 | 0.81 | 0.038 |
| Gene Name | Gene ID | p (corr) Value | Fold Change | GO Biological Process |
|---|---|---|---|---|
| Agpat9 | 1-Acylglycerol-3-phosphate-o-acyltransferase 9 | 8.10 × 10−4 | 3.740 | Lipid metabolic process |
| Cdkn1a | Cyclin-dependent kinase inhibitor1A (P21) | 4.24 × 10−4 | 3.153 | Regulation of cyclin-dependent protein serine |
| Elovl3 | Elongation of very long chain fatty acids-like3 | 3.76 × 10−3 | 3.055 | Lipid metabolic process |
| Pnpla2 | Patatin-like phospholipase domain containing 2 | 1.17 × 10−3 | 2.645 | Lipid metabolic process |
| Pde4d | Phosphodiesterase 4D, cAMP specific | 7.68 × 10−4 | 2.161 | cAMP catabolic process |
| Pex11a | Peroxisomal biogenesis factor 11 alpha | 1.34 × 10−3 | 2.066 | Peroxisome organization |
| Pex3 | Peroxisomal biogenesis factor 3 | 8.82 × 10−3 | 2.037 | Peroxisome organization |
| Pex19 | Peroxisomal biogenesis factor 19 | 3.23 × 10−3 | 1.953 | Protein targeting to peroxisome |
| Cirbp | Cold inducible RNA binding protein | 1.15 × 10−2 | 1.895 | Response to stress |
| Pex16 | Peroxisomal biogenesis factor 16 | 4.73 × 10−4 | 1.887 | Protein targeting to peroxisome |
| Acot8 | Acyl-Coa-thioesterase 8 | 4.73 × 10−4 | 1.776 | Peroxisome organization |
| Atg2a | Autophagy related 2A | 2.59 × 10−3 | 1.707 | Autophagy |
| Vwa8 | Von willebrand factor A domain containing 8 | 1.94 × 10−3 | 1.651 | ATP catabolic process |
| Abcd3 | ATP-binding cassette, sub-familyD (ALD), member 3 | 1.94 × 10−3 | 1.651 | ATP catabolic process |
| Abcg8 | ATP-binding cassette, sub-family G (WHITE), member 8 | 2.08 × 10−2 | 1.628 | ATP catabolic process |
| Atp10d | ATPase, class V, type 10D | 7.48 × 10−3 | 1.614 | ATP catabolic process |
| Map1lc3b | Microtubule-associated protein 1 light chain 3 β | 2.50 × 10−2 | 1.553 | Autophagy |
| Abcg5 | ATP binding cassette subfamily G member 5 | 4.11 × 10−2 | 1.550 | ATP catabolic process |
| Ppara | peroxisome proliferator activated receptor α | 2.10 × 10−3 | 1.550 | Negative regulation of transcription |
| Mtor | Mechanistic target of rapamycin | 5.06 × 10−3 | 1.541 | Positive regulation of protein phosphorylation |
| Wipi1 | WD repeat domain, phosphoinositide interacting 1 | 3.81 × 10−2 | 1.537 | Autophagic vacuole assembly |
| Ppargc1b | PPARG coactivator 1 β | 1.24 × 10−2 | 1.535 | Transcription from mitochondrial promoter |
| Atg14 | Autophagy related 14 | 2.54 × 10−2 | 1.505 | Autophagic vacuole assembly |
| Pex1 | Peroxisomal biogenesis factor 1 | 8.32 × 10−3 | 1.417 | Protein targeting to peroxisome |
| Pex11b | Peroxisomal biogenesis factor | 2.49 × 10−2 | 1.365 | Peroxisome organization |
| Pex10 | Peroxisomal biogenesis factor 10 | 3.85 × 10−2 | 1.357 | Peroxisome organization |
| Wipi2 | WD repeat domain, phosphoinositide interacting 2 | 1.67 × 10−2 | 1.277 | Autophagic vacuole assembly |
| Map1lc3a | Microtubule associated protein 1 light chain 3 α | 1.30 × 10−2 | 1.256 | Autophagic vacuole assembly |
| Ampd2 | Adenosine monophosphate deaminase 2 | 1.01 × 10−2 | −1.608 | AMP biosynthetic process |
| Nfe2l2 | Nuclear factor, erythroid 2 like 2 | 5.80 × 10−3 | −1.643 | Transcription, DNA-dependent |
| Cyp26a1 | Cytochrome P450 family 26 subfamily A member 1 | 4.97 × 10−2 | −2.015 | Central nervous system development |
| Hsp90aa1 | Heat shock protein 90 α family class A member 1 | 7.68 × 10−4 | −2.653 | ATP catabolic process |
| Hspb1 | Heat shock protein family B (small) member 1 | 1.03 × 10−3 | −5.755 | Response to stress |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blaber, E.A.; Pecaut, M.J.; Jonscher, K.R. Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver. Int. J. Mol. Sci. 2017, 18, 2062. https://doi.org/10.3390/ijms18102062
Blaber EA, Pecaut MJ, Jonscher KR. Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver. International Journal of Molecular Sciences. 2017; 18(10):2062. https://doi.org/10.3390/ijms18102062
Chicago/Turabian StyleBlaber, Elizabeth A., Michael J. Pecaut, and Karen R. Jonscher. 2017. "Spaceflight Activates Autophagy Programs and the Proteasome in Mouse Liver" International Journal of Molecular Sciences 18, no. 10: 2062. https://doi.org/10.3390/ijms18102062






