Hydrogen Peroxide Response in Leaves of Poplar (Populus simonii × Populus nigra) Revealed from Physiological and Proteomic Analyses
Abstract
:1. Introduction
2. Results
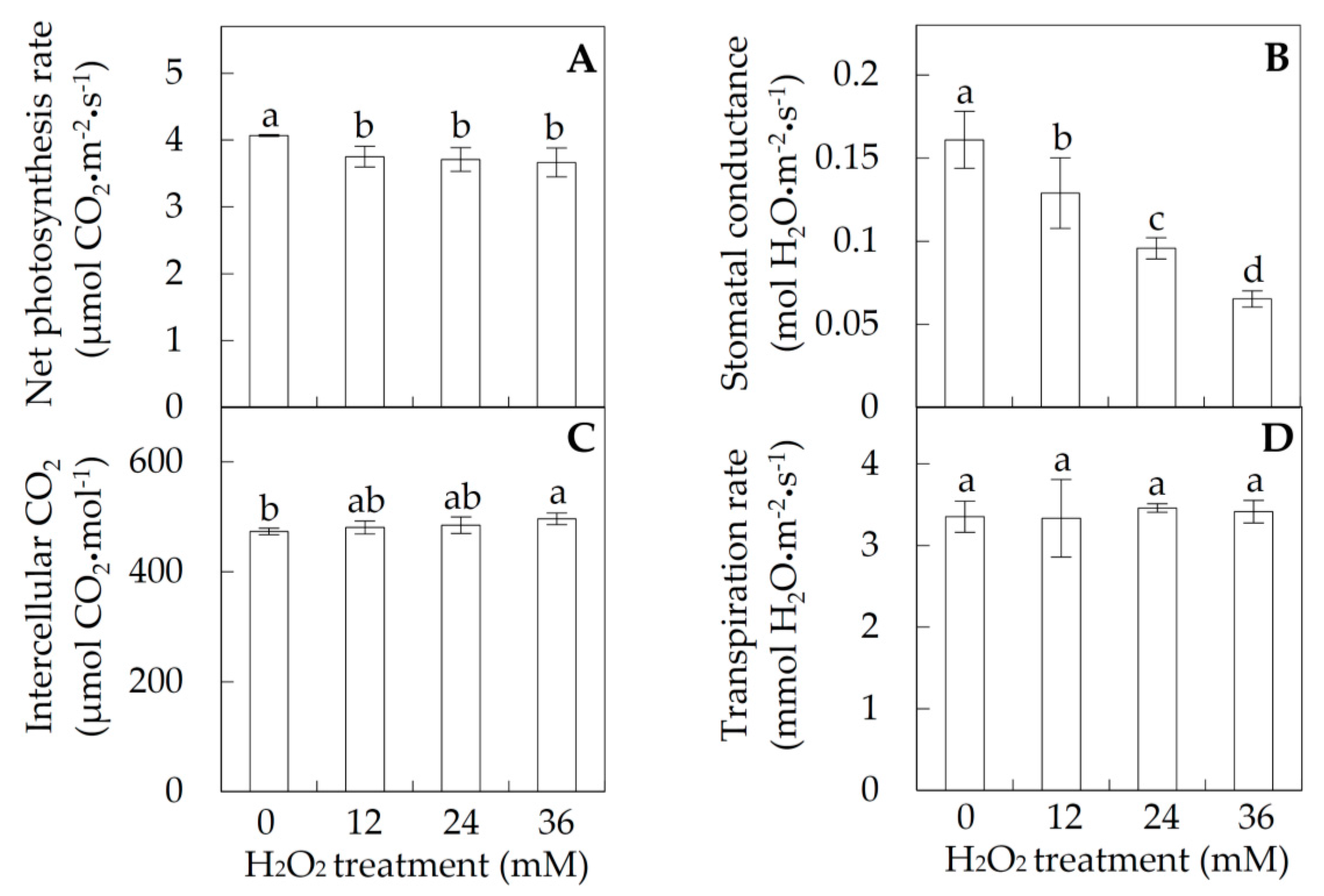
2.1. Photosynthesis under Hydrogen Peroxide (H2O2) Stress
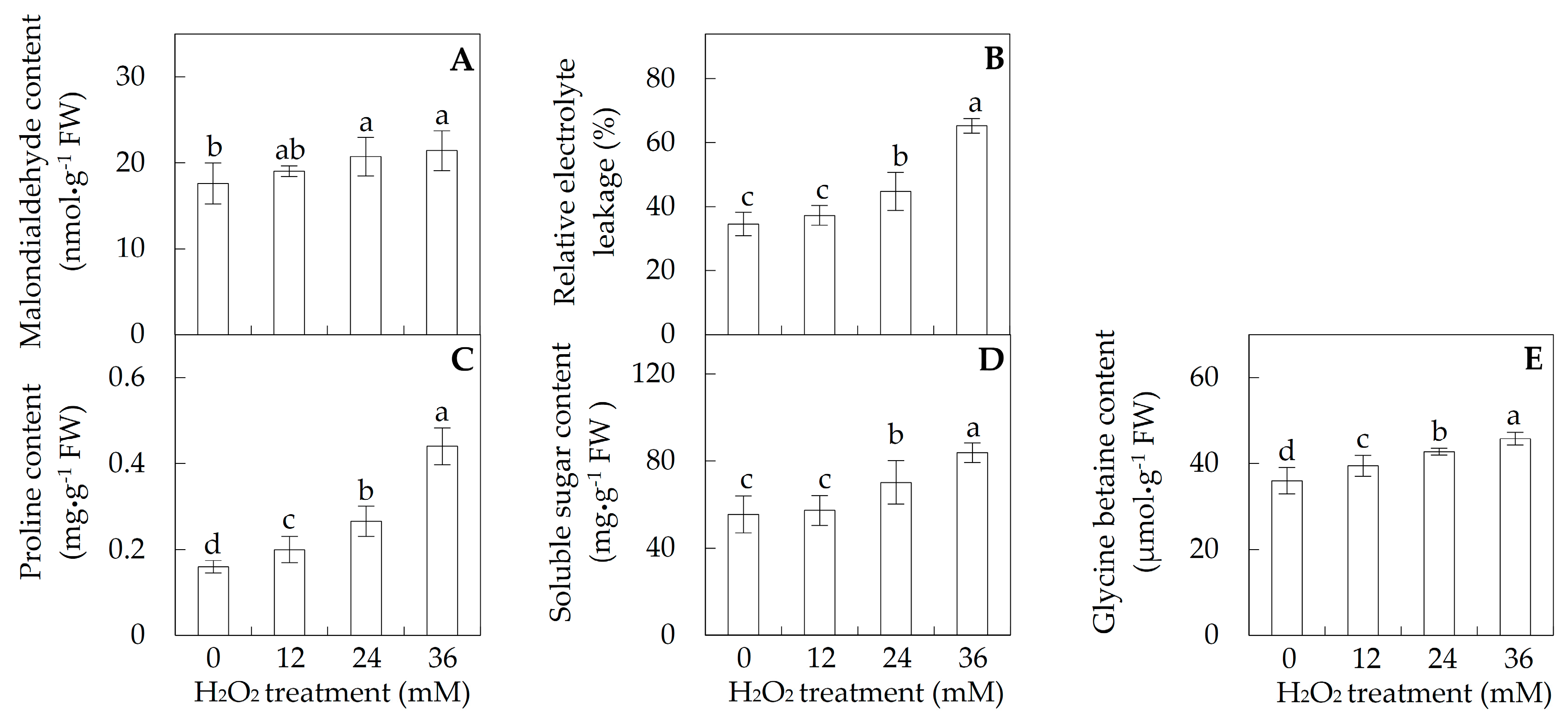
2.2. Membrane Integrity and Osmolyte Accumulation in Leaves
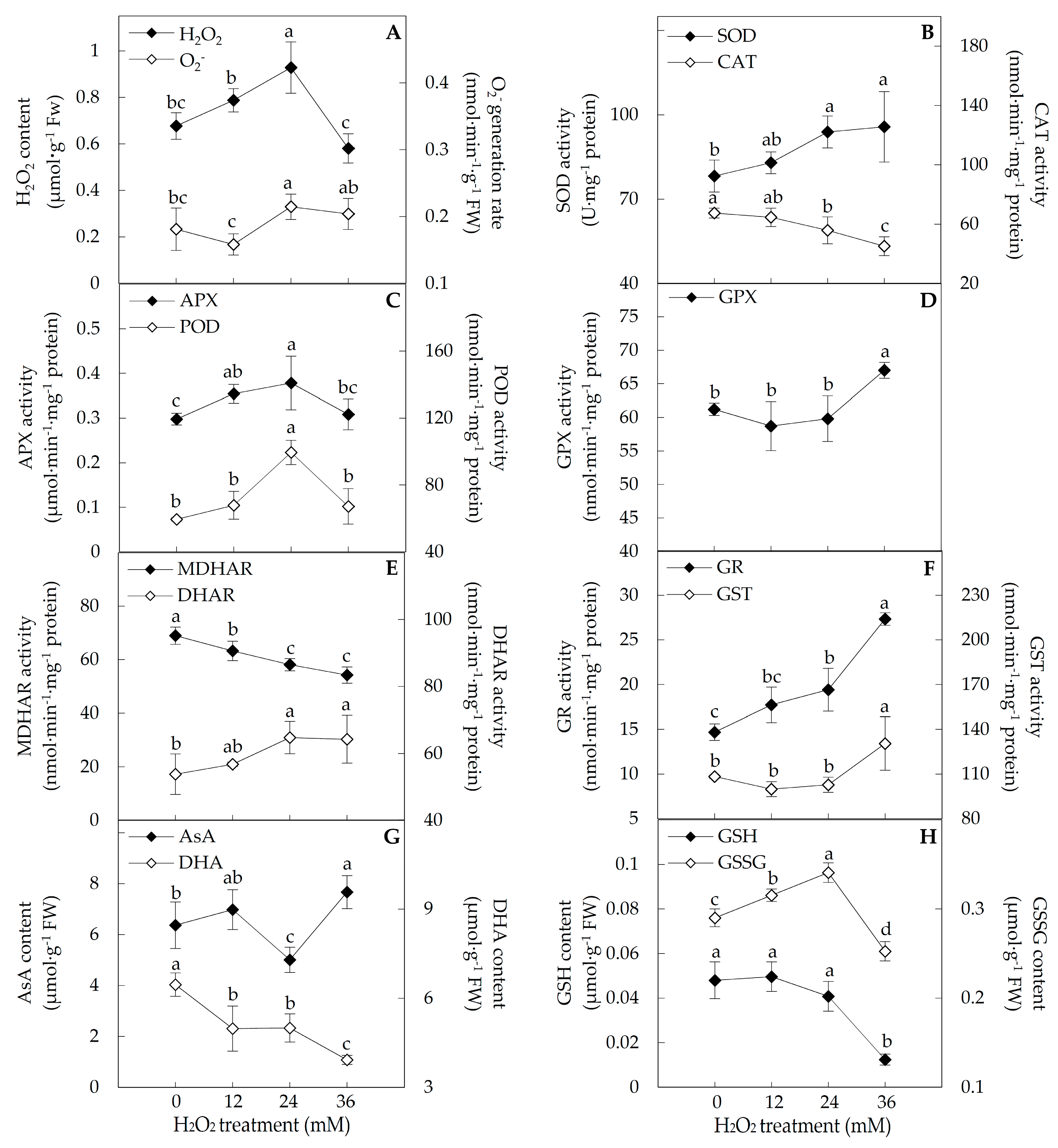
2.3. ROS and Antioxidant Substances Content, and Antioxidant Enzyme Activities
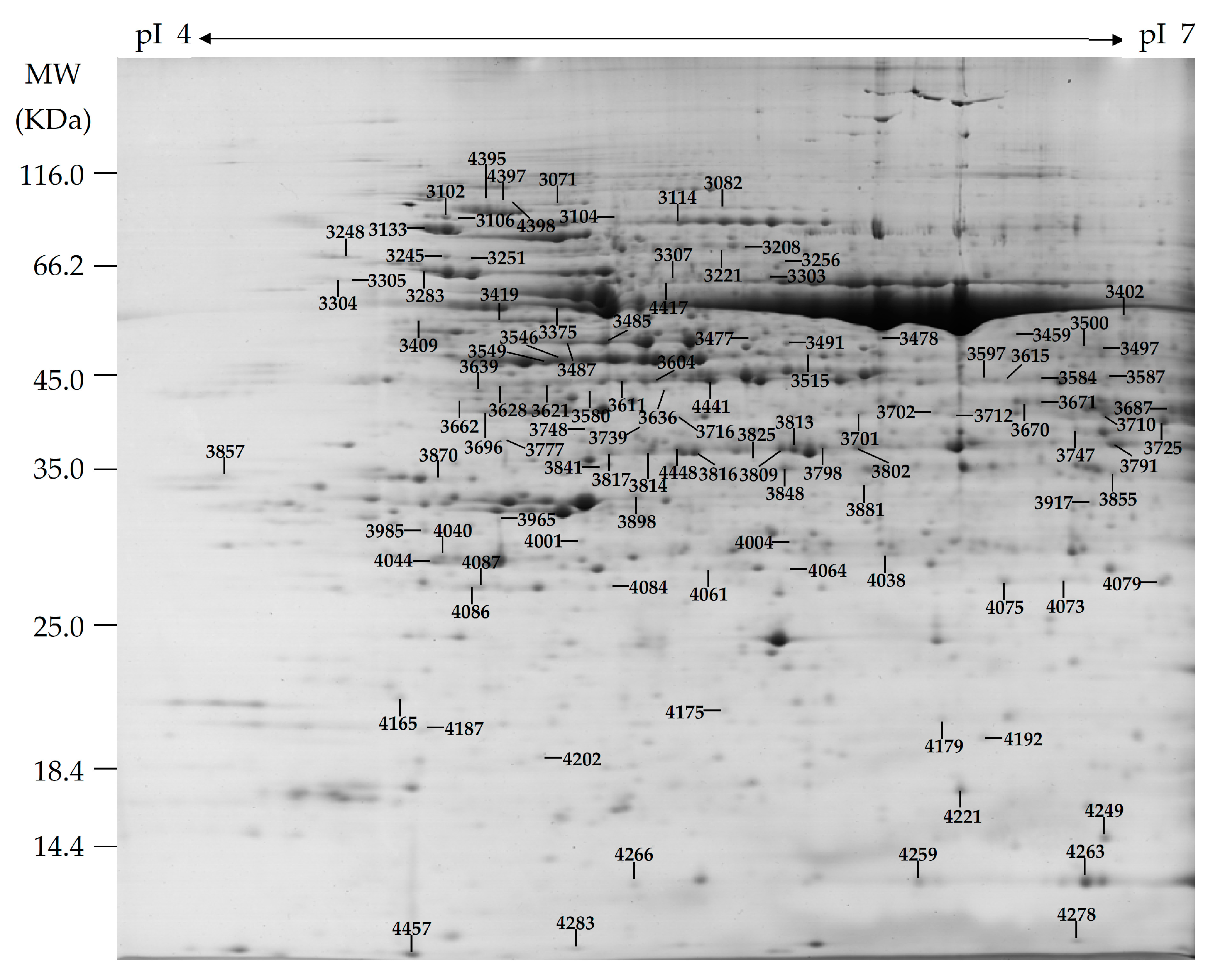
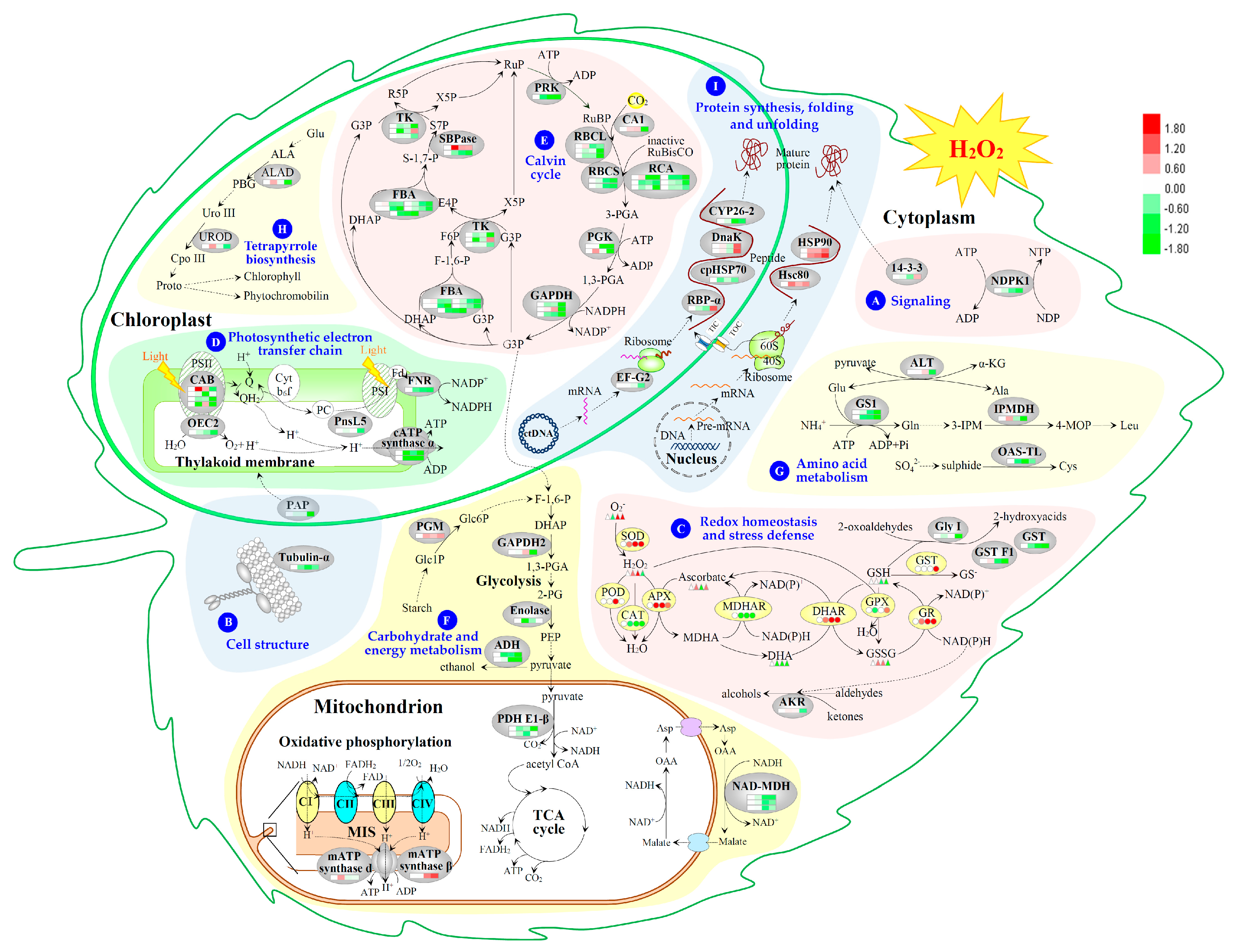
2.4. Identification of H2O2-Responsive Proteins in Leaves
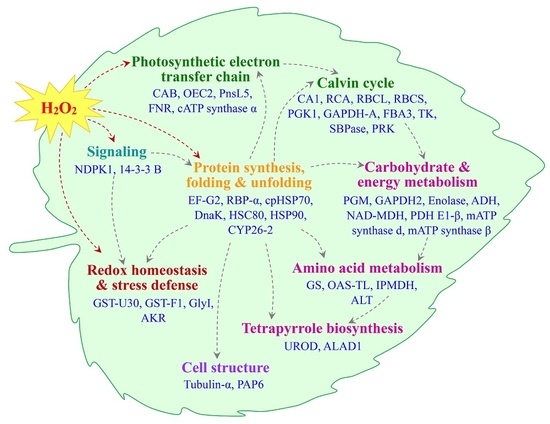
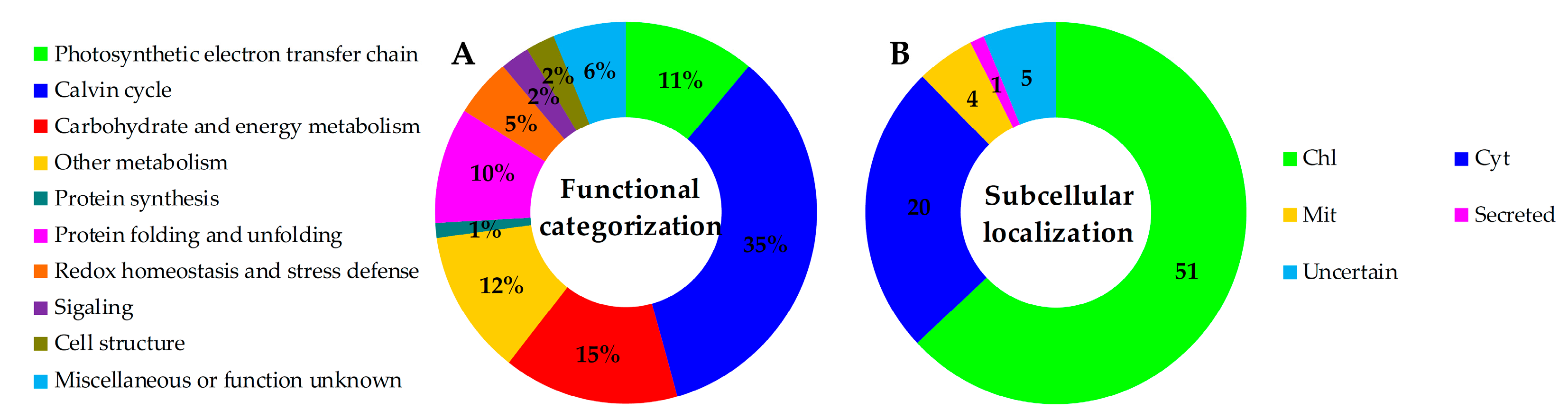
2.5. Annotation and Functional Categorization of the H2O2-Responsive Proteins
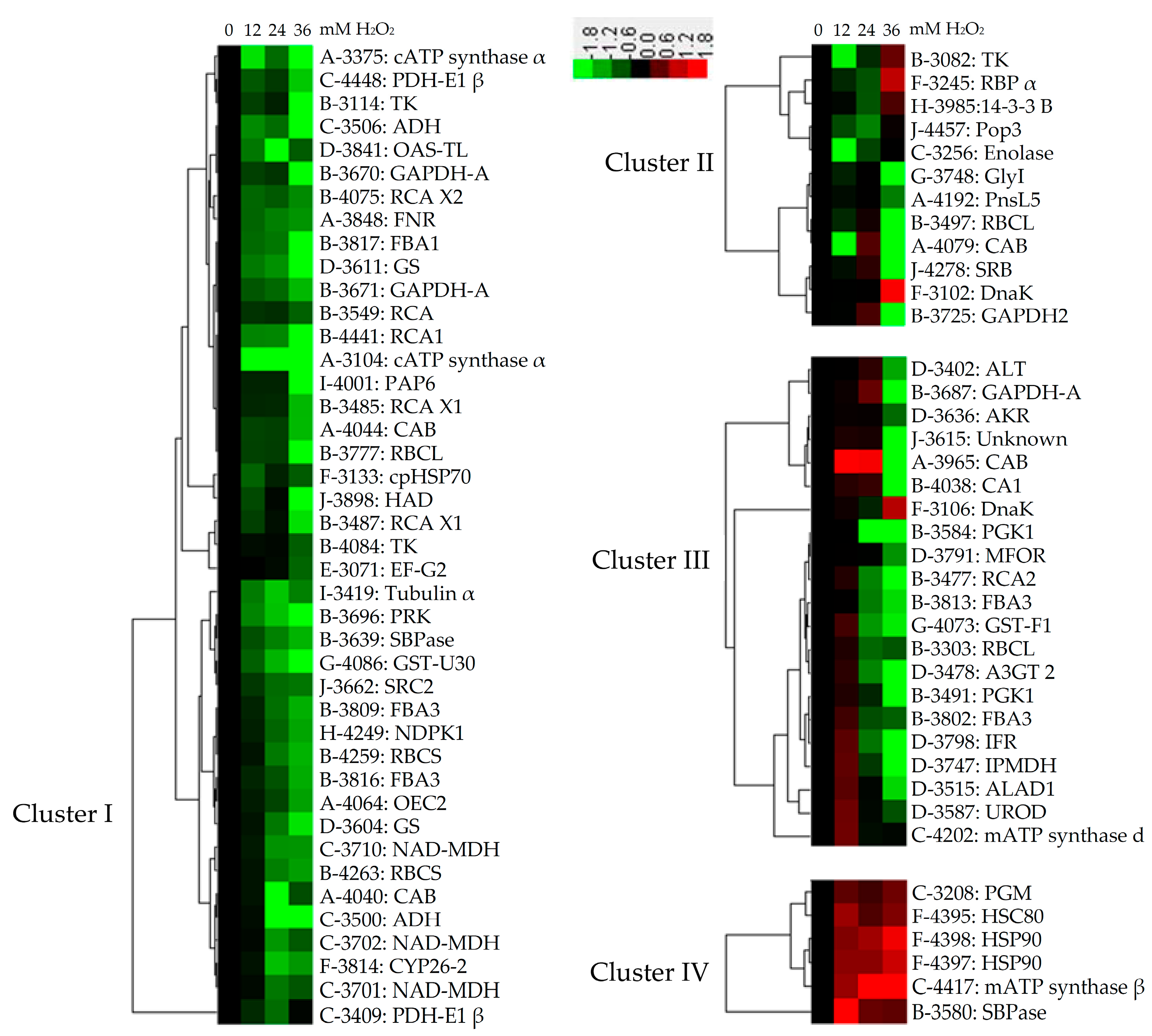
2.6. Subcellular Localization and Hierarchical Clustering of H2O2-Responsive Proteins
2.7. Hierarchical Clustering and Analysis of H2O2-Responsive Proteins
2.8. Protein–Protein Interaction (PPI) among H2O2-Responsive Proteins
3. Discussion
3.1. H2O2-Responsive Signal Transduction and Cellular Structure Modulation in Poplar
3.2. H2O2-Induced Alteration of ROS Scavenging Pathways in Poplar
3.3. H2O2-Altered Redox Homeostasis in Poplar Leaves
3.4. H2O2-Reduced Photosynthesis in Poplar
3.5. H2O2-Responsive Carbohydrate and Other Metabolisms in Poplar
3.6. H2O2-Responsive Proteins Conformation in Poplar
4. Methods
4.1. Plant Cultivation and Treatment
4.2. Photosynthesis Measurement
4.3. Determination of MDA Content, REL, Total Soluble Sugar, Proline, and Glycine Betaine Contents
4.4. Determination of ROS and Antioxidant Substances Contents, and Antioxidant Enzyme Activity Assay
4.5. Protein Sample Preparation, 2DE, and Protein Abundance Analysis
4.6. Protein Identification by MALDI-TOF/TOF MS and Database Searching
4.7. Protein Classification, Subcellular Localization, Hierarchical Cluster Analysis, and Protein–Protein Interaction Prediction
4.8. Statistical Analysis
5. Conclusion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 14-3-3 B | 14-3-3-like protein B |
| 2DE | Two-dimensional gel electrophoresis |
| 1O2 | Singlet oxygen |
| A3GT 2 | Anthocyanidin 3-O-glucosyltransferase 2 |
| ADH | Alcohol dehydrogenase |
| AKR | Aldo/keto reductase family protein |
| ALAD1 | Delta-aminolevulinic acid dehydratase 1 |
| ALT | Alanine aminotransferase family protein |
| APX | Ascorbate peroxidase |
| AsA | Ascorbate |
| BLAST | Basic local alignment search tool |
| CA1 | Carbonic anhydrase isoform 1 |
| CAB | Chlorophyll A/B binding protein |
| CAT | Catalase |
| cATP synthase α | ATP synthase CF1 α subunit |
| CBB | Coomassie brilliant blue |
| Ci | Intercellular CO2 concentration |
| cpHSP70 | Stromal 70 kda heat shock-related family protein |
| CYP26-2 | Peptidyl-prolyl cis-trans isomerase CYP26-2 |
| DAP | Differentially accumulated protein |
| DHA | Dehydroascorbate |
| DHAR | Dehydroascorbate reductase |
| DnaK | Chaperone Dnak |
| EF-G2 | Elongation factor G-2 |
| ESTs | Expressed sequence tags |
| FBA | Fructose-bisphosphate aldolase |
| FNR | Ferredoxin-NADP reductase |
| GAPDH2 | Glyceraldehyde-3-phosphate dehydrogenase 2 |
| GAPDH-A | Glyceraldehyde-3-phosphate dehydrogenase A |
| Gly I | Glyoxalase I homolog family protein |
| GPX | Glutathione peroxidase |
| GR | Glutathione reductase |
| GS | Glutamine synthetase |
| Gs | Stomatal conductance |
| GSH | Reduced glutathione |
| GSSG | Oxidized glutathione |
| GST | Glutathione S-transferase |
| GST-F1 | Glutathione S-transferase F1 (GST-F1) |
| GST-U30 | Glutathione S-transferase U30 |
| H2O2 | Hydrogen peroxide |
| HAD | Haloacid dehalogenase-like hydrolase family protein |
| HO• | Hydroxyl radicals |
| HSC80 | Heat shock cognate protein 80 |
| HSP90 | Heat shock protein 90 |
| IFR | Isoflavone reductase family protein |
| IPG | Immobilized pH gradient |
| IPMDH | 3-isopropyl malate dehydrogenase |
| iTRAQ | Isobaric tags for relative and absolute quantification |
| mATP synthase d | ATP synthase subunit d |
| mATP synthase β | ATP synthase subunit beta |
| MDA | Malondialdehyde |
| MDHAR | Monodehydroascorbate reductase |
| MFOR | 2-methylene-furan-3-one reductase |
| MALDI | Matrix-assisted laser desorption/ ionization |
| MS | Mass spectrometry |
| NAD-MDH | NAD-dependent malate dehydrogenase |
| NADH | Nicotinamide adenine dinucleotide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NDPK1 | Nucleoside diphosphate kinase 1 |
| O2•− | Superoxide anion radicals |
| OAS-TL | O-acetylserine (thiol) lyase family protein |
| OEC2 | Photosystem II oxygen-evolving complex protein 2 precursor |
| PAP6 | Plastid-lipid-associated protein 6 |
| PDH E1-β | Pyruvate dehydrogenase E1 component subunit beta |
| PGK1 | Phosphoglycerate Kinase 1 |
| PGM | Phosphoglucomutase |
| Pn | Net photosynthetic rate |
| PnsL5 | Photosynthetic NDH subunit of lumenal location 5 |
| POD | Peroxidase |
| Pop3 | Pop3 peptide family protein |
| PRK | Phosphoribulokinase |
| PTM | post-translational modification |
| RBCL | Rubisco large chain |
| RBCS | Rubisco small chain |
| RBP-α | Rubisco large subunit-binding protein subunit α |
| RCA | Rubisco activase |
| REL | Relative electrolyte leakage |
| ROS | Reactive oxygen species |
| RuBisCO | Ribulose-bisphosphate carboxylase |
| SBPase | Sedoheptulose-1, 7-bisphosphatase |
| SOD | Superoxide dismutase |
| SRB | Stress responsive A/B Barrel domain |
| SRC2 | Soybean genes regulated by cold 2 (SRC2)-like domain |
| TCA | Tricarboxylic acid |
| TK | Transketolase |
| TOF | Time of flight |
| Tr | Transpiration rate |
| Tubulin α | Tubulin α chain |
| UROD | Uroporphyrinogen decarboxylase |
References
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van, B.F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R. ROS are good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Petrov, V.D.; van Breusegem, F. Hydrogen peroxide—A central hub for information flow in plant cells. AoB Plants 2012, 2012. [Google Scholar] [CrossRef] [PubMed]
- Rinalducci, S.; Murgiano, L.; Zolla, L. Redox proteomics: Basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 2008, 59, 3781–3801. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, Y.; Yamamoto, K.; Tawaratsumida, T.; Yuasa, T.; Iwaya-Inoue, M. Hydrogen peroxide scavenging regulates germination ability during wheat (Triticum aestivum L.) seed maturation. Plant Sign. Behav. 2008, 3, 183–188. [Google Scholar] [CrossRef]
- Uchida, A.; Jagendorf, A.T.; Hibino, T.; Takabe, T.; Takabe, T. Effects of hydrogen peroxide and nitric oxide on both salt and heat stress tolerance in rice. Plant Sci. 2002, 163, 515–523. [Google Scholar] [CrossRef]
- Rokebul Anower, M.; Peel, M.D.; Mott, I.W.; Wu, Y. Physiological processes associated with salinity tolerance in an alfalfa half-sib family. J. Agro. Crop Sci. 2017. [Google Scholar] [CrossRef]
- Liu, Z.J.; Guo, Y.K.; Bai, J.G. Exogenous hydrogen peroxide changes antioxidant enzyme activity and protects ultrastructure in leaves of two cucumber ecotypes under osmotic stress. J. Plant Growth Regul. 2010, 29, 171–183. [Google Scholar] [CrossRef]
- Xu, F.J.; Jin, C.W.; Liu, W.J.; Zhang, Y.S.; Lin, X.Y. Pretreatment with H2O2 alleviates aluminum-induced oxidative stress in wheat seedlings. J. Integr. Plant Biol. 2011, 53, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Sirhindi, G.; Bhardwaj, R.; Kumar, S.; Jain, G. Effect of exogenous H2O2 on antioxidant enzymes of Brassica juncea L. seedlings in relation to 24-epibrassinolide under chilling stress. Indian J. Biochem. Biophys. 2010, 47, 378–382. [Google Scholar] [PubMed]
- Moskova, I.; Todorova, D.; Alexieva, V.; Segiev, I. Protective effect of hydrogen peroxide against paraquat toxicity in young pea plants: Possible role of endogenous polyamines. Am. J. Plant Sci. 2014, 5, 3408–3416. [Google Scholar] [CrossRef]
- Bădărău, C.L.; Chiru, N.; Damsa, F.; Nistor, A. Effects of Satureja hortensis oil treatments and exogenous H2O2 on Potato virus Y (PVY) infected Solanum tuberosum L. plants under drought conditions. An. Univers. Orad. Fasc. Biol. 2012, 19, 140–145. [Google Scholar]
- Desikan, R.; A-H-Mackerness, S.; Hancock, J.T.; Neill, S.J. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001, 127, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, S.; van der Kelen, K.; Dat, J.; Gadjev, I.; Boonefaes, T.; Morsa, S.; Rottiers, P.; Slooten, L.; van Montagu, M.; Zabeau, M.; et al. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proc. Natl. Acad. Sci. USA 2003, 100, 16113–16118. [Google Scholar] [CrossRef] [PubMed]
- Vanderauwera, S.; Zimmermann, P.; Rombauts, S.; Vandenabeele, S.; Langebartels, C.; Gruissem, W.; Inze, D.; van Breusegem, F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005, 139, 806–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.L.; Zhang, R.Z.; Pan, L.; Tang, L.C.; Zhao, G.G.; Zhu, M.Z.; Chu, J.F.; Sun, X.H.; Wei, B.; Zhang, X.Q.; et al. Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PLoS ONE 2011, 6, e28810. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.Y.; Liu, J.Y. Comparative proteomics analysis reveals an intimate protein network provoked by hydrogen peroxide stress in rice seedling leaves. Mol. Cell. Proteom. 2008, 7, 1469–1488. [Google Scholar] [CrossRef] [PubMed]
- Tanou, G.; Job, C.; Belghazi, M.; Molassiotis, A.; Diamantidis, G.; Job, D. Proteomic signatures uncover hydrogen peroxide and nitric oxide cross-talk signaling network in citrus plants. J. Proteome Res. 2010, 9, 5994–6006. [Google Scholar] [CrossRef] [PubMed]
- Bian, Y.W.; Lv, D.W.; Cheng, Z.W.; Gu, A.Q.; Cao, H.; Yan, Y.M. Integrative proteome analysis of Brachypodium distachyon roots and leaves reveals a synergetic responsive network under H2O2 stress. J. Proteom. 2015, 128, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Ge, P.; Hao, P.; Cao, M.; Guo, G.; Lv, D.; Subburaj, S.; Li, X.; Yan, X.; Xiao, J.; Ma, W.; et al. iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways of wheat seedling growth under hydrogen peroxide stress. Proteomics 2013, 13, 3046–3058. [Google Scholar] [CrossRef] [PubMed]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.Q.; Liu, X.; Jiang, Z.H.; Wang, Y.h.; Yu, H.Q. Effects of planting density on wood anatomical properties of Populus × xiaohei. For. Res. 2006, 19, 364–369. [Google Scholar]
- Gao, W.; Bai, S.; Li, Q.; Gao, C.; Liu, G.; Li, G.; Tan, F. Overexpression of TaLEA gene from Tamarix androssowii improves salt and drought tolerance in transgenic poplar (Populus simonii × P. nigra). PLoS ONE 2013, 8, e67462. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.W.; Liu, G.F.; Wang, Z.Y.; Yan, S.C.; Ma, L.; Yang, C.P. Response of the gypsy moth, Lymantria dispar to transgenic poplar, Populus simonii × P. nigra, expressing fusion protein gene of the spider insecticidal peptide and Bt-toxin C-peptide. J. Insect Sci. 2010, 10, 1–13. [Google Scholar] [CrossRef]
- Wang, Z.B. Study on poplar transgene of the fungus disease-resistance. Forest. Sci. Tech. 2006, 31, 22–24. [Google Scholar]
- Wang, L.; Zhou, B.; Wu, L.L.; Guo, B.Z.; Jiang, T.B. Differentially expressed genes in Populus simonii × Populus nigra in response to NaCl stress using cDNA-AFLP. Plant Sci. 2011, 180, 796–801. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jiang, J.; Li, H.; Liu, G. The salt-responsive transcriptome of Populus simonii × Populus nigra via DGE. Gene 2012, 504, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.M.; Chen, S.; Lin, L.; Wei, R.; Li, H.Y.; Liu, G.F.; Jiang, J. Genome-wide analysis of a TaLEA-introduced transgenic Populus simonii × Populus nigra dwarf mutant. Int. J. Mol. Sci. 2012, 13, 2744–2762. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yuan, H.M.; Liu, G.F.; Li, H.Y.; Jiang, J. A label-free differential quantitative proteomics analysis of a TaLEA-introduced transgenic Populus simonii × Populus nigra dwarf mutant. Mol. Biol. Rep. 2012, 39, 7657–7664. [Google Scholar] [CrossRef] [PubMed]
- STRING: Functional Protein Association Networks. Available online: http://string-db.org (accessed on 2 April 2017).
- TAIR BLAST 2.2.8. Available online: http:// www.arabidopsis.org/Blast/index.jsp (accessed on 2 April 2017).
- Chang, I.F.; Curran, A.; Woolsey, R.; Quilici, D.; Cushman, J.C.; Mittler, R.; Harmon, A.; Harper, J.F. Proteomic profiling of tandem affinity purified 14-3-3 protein complexes in Arabidopsis thaliana. Proteomics 2009, 9, 2967–2985. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.R.; Salinas, J.; Collinge, D.B. 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 2002, 50, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Lee, B.; Choi, G.; Shin, D.; Prasad, D.T.; Lee, O.; Kwak, S.S.; Kim, D.H.; Nam, J.; Bahk, J.; et al. NDP kinase 2 interacts with two oxidative stress-activated MAPKs to regulate cellular redox state and enhances multiple stress tolerance in transgenic plants. Proc. Natl. Acad. Sci. USA 2003, 100, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Dorion, S.; Rivoal, J. Clues to the functions of plant NDPK isoforms. Naunyn-Schmiedeberg's Arch. Pharmacol. 2015, 388, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Leitnerdagan, Y.; Ovadis, M.; Shklarman, E.; Elad, Y.; Rav, D.D.; Vainstein, A. Expression and functional analyses of the plastid lipid-associated protein CHRC suggest its role in chromoplastogenesis and stress. Plant Physiol. 2006, 142, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Gillet, B.; Beyly, A.; Peltier, G.; Rey, P. Molecular characterization of CDSP 34, a chloroplastic protein induced by water deficit in Solanum tuberosum L. plants, and regulation of CDSP 34 expression by ABA and high illumination. Plant J. 1998, 16, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.U.; Wu, S.S.; Ratnayake, C.; Huang, A.H. Brassica rapa has three genes that encode proteins associated with different neutral lipids in plastids of specific tissues. Plant Physiol. 2001, 126, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Laizet, Y.; Pontier, D.; Mache, R.; Kuntz, M. Subfamily organization and phylogenetic origin of genes encoding plastid lipid-associated proteins of the fibrillin type. J. Genome Technol. Sci. 2004, 3, 19–28. [Google Scholar] [CrossRef]
- Bartolo, M.E.; Carter, J.V. Microtubules in mesophyll cells of nonacclimated and cold-acclimated spinach: Visualization and responses to freezing, low temperature, and dehydration. Plant Physiol. 1991, 97, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Abdrakhamanova, A.; Wang, Q.Y.; Khokhlova, L.; Nick, P. Is microtubule disassembly a trigger for cold acclimation? Plant Cell Physiol. 2003, 44, 676–686. [Google Scholar] [CrossRef] [PubMed]
- Shoji, T.; Suzuki, K.; Abe, T.; Kaneko, Y.; Shi, H.; Zhu, J.K.; Rus, A.; Hasegawa, P.M.; Hashimoto, T. Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant Cell Physiol. 2006, 47, 1158–1168. [Google Scholar] [CrossRef] [PubMed]
- Ban, Y.; Kobayashi, Y.; Hara, T.; Hamada, T.; Hashimoto, T.; Takeda, S.; Hattori, T. α-Tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol. 2013, 54, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.L. Effects of H2O2 stress on physiological and biochemical index of Grass Pea. Acta Agr. Boreali-Occident. Sin. 2015, 25, 243–248. [Google Scholar] [CrossRef]
- Yoshimura, K.; Miyao, K.; Gaber, A.; Takeda, T.; Kanaboshi, H.; Miyasaka, H.; Shigeoka, S. Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 2004, 37, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Sytykiewicz, H. Expression patterns of genes involved in ascorbate-glutathione cycle in aphid-infested maize (Zea mays L.) seedlings. Int. J. Mol. Sci. 2016, 17, 268. [Google Scholar] [CrossRef] [PubMed]
- Mohsenzadeh, S.; Esmaeili, M.; Moosavi, F.; Shahrtash, M.; Saffari, B.; Mohabatkar, H. Plant glutathione S-transferase classification, structure and evolution. Afr. J. Biotechnol. 2011, 10, 8160–8165. [Google Scholar]
- Kaur, C.; Singlapareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Sengupta, D.; Naik, D.; Reddy, A.R. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. J. Plant Physiol. 2015, 179, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Zhang, H.; Wang, H.; Xia, Y. Identification of redox-sensitive cysteines in the Arabidopsis proteome using OxiTRAQ, a quantitative redox proteomics method. Proteomics 2014, 14, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Guo, G.; Yang, Z.; Li, Y.; Xia, Y.; Li, N. Stable isotope metabolic labeling-based quantitative thiol redox proteomic analysis of hydrogen peroxide-treated Arabidopsis plant. J. Proteom. Bioinformat. 2014, 7, 121–133. [Google Scholar] [CrossRef]
- Muthuramalingam, M.; Matros, A.; Scheibe, R.; Mock, H.P.; Dietz, K.J. The hydrogen peroxide-sensitive proteome of the chloroplast in vitro and in vivo. Front. Plant Sci. 2013, 4, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, S.; Lu, Y.; Alvarez, S.; Hicks, L.M.; Ge, X.; Xia, Y. Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis. J. Proteome Res. 2012, 11, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Regier, N.; Streb, S.; Cocozza, C.; Schaub, M.; Cherubini, P.; Zeeman, S.C.; Frey, B. Drought tolerance of two black poplar (Populus nigra L.) clones: Contribution of carbohydrates and oxidative stress defence. Plant Cell Environ. 2009, 32, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.H.; Dennis, E.S.; Peacock, W.J. Arabidopsis roots and shoots have different mechanisms for hypoxic stress tolerance. Plant Physiol. 1999, 119, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Musrati, R.A.; Kollárová, M.; Mernik, N.; Mikulásová, D. Malate dehydrogenase: Distribution, function and properties. Gen. Physiol. Biophys. 1998, 17, 193–210. [Google Scholar] [PubMed]
- Tischner, R. Nitrate uptake and reduction in higher and lower plants. Plant Cell Environ. 2000, 23, 1005–1024. [Google Scholar] [CrossRef]
- Zagorchev, L.; Seal, C.E.; Kranner, I.; Odjakova, M. A central role for thiols in plant tolerance to abiotic stress. Int. J. Mol. Sci. 2013, 14, 7405–7432. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.B.; Tabaldi, L.A.; Gonçalves, J.F.; Jucoski, G.O.; Pauletto, M.M.; Weis, S.N.; Nicoloso, F.T.; Borher, D.; Rocha, J.B.T.; Schetinger, M.R.C. Effect of aluminum on δ-aminolevulinic acid dehydratase (ALA-D) and the development of cucumber (Cucumis sativus). Environ. Exp. Bot. 2006, 57, 106–115. [Google Scholar] [CrossRef]
- Skrebsky, E.C.; Tabaldi, L.A.; Pereira, L.B.; Rauber, R.; Maldaner, J.; Cargnelutti, D.; Gonçalves, J.F.; Castro, G.Y.; Shetinger, M.R.; Nicoloso, F.T. Effect of cadmium on growth, micronutrient concentration, and δ-aminolevulinic acid dehydratase and acid phosphatase activities in plants of Pfaffia glomerata. Braz. J. Plant Physiol. 2008, 20, 285–294. [Google Scholar] [CrossRef]
- Gupta, D.K.; Nicoloso, F.T.; Schetinger, M.R.; Rossato, L.V.; Pereira, L.B.; Castro, G.Y.; Srivastava, S.; Tripathi, R.D. Antioxidant defense mechanism in hydroponically grown Zea mays seedlings under moderate lead stress. J. Hazard. Mater. 2009, 172, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Mock, H.P.; Keetman, U.; Kruse, E.; Rank, B.; Grimm, B. Defense responses to tetrapyrrole-induced oxidative stress in transgenic plants with reduced uroporphyrinogen decarboxylase or coproporphyrinogen oxidase activity. Plant Physiol. 1998, 116, 107–116. [Google Scholar] [CrossRef]
- Wang, W.X.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Demirevska, K.; Simova-Stoilova, L.; Vassileva, V.; Feller, U. Rubisco and some chaperone protein responses to water stress and rewatering at early seedling growth of drought sensitive and tolerant wheat varieties. Plant Growth Regul. 2008, 56, 97–106. [Google Scholar] [CrossRef]
- Caruso, G.; Cavaliere, C.; Guarino, C.; Gubbiotti, R.; Foglia, P.; Laganà, A. Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal. Bioanal. Chem. 2008, 391, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, M.E. Association of Rubisco activase with chaperonin-60β: A possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 2008, 59, 1923–1933. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.M.; Li, P.; Tang, T.; Wang, J.C.; Chen, Y.; Liu, L. Roles of Hsp70s in stress responses of microorganisms, plants, and animals. BioMed Res. Int. 2015, 2015, 510319. [Google Scholar] [CrossRef] [PubMed]
- Schroda, M.; Vallon, O.; Wollman, F.-A.; Beck, C.F. A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell 1999, 11, 1165–1178. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.H.; Guo, J.; Ding, K.; Wang, S.J.; Zhang, H.; Dai, X.W.; Chen, Y.Y.; Govers, F.; Huang, L.L.; Kang, Z.S. Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol. Biol. Rep. 2011, 38, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Scharf, K.D. Synthesis, modification and structural binding of heat-shock proteins in tomato cell cultures. Eur. J. Biochem. 1984, 139, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Li, Z.Y.; Chen, Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Heat shock protein 90 in plants: Molecular mechanisms and roles in stress responses. Int. J. Mol. Sci. 2012, 13, 15706–15723. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, S.; Zhao, Q.; Wang, T.; Yang, C.; Diaz, C.; Sun, G.; Dai, S. Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J. Proteome Res. 2011, 10, 3852–3870. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, S.; Zhang, H.; Shi, L.; Cao, F.; Guo, L.; Xie, Y.; Wang, T.; Yan, X.; Dai, S. Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J. Proteome Res. 2010, 9, 6561–6577. [Google Scholar] [CrossRef] [PubMed]
- Li, H.S.; Sun, Q.; Zhao, S.J.; Zhang, W.H. Principles and Techniques of Plant Physiological Biochemical Experiment; Higher Education Press: Beijing, China, 2000; pp. 186–191. [Google Scholar]
- Zhao, Q.; Suo, J.; Chen, S.; Jin, Y.; Ma, X.; Yin, Z.; Zhang, Y.; Wang, T.; Luo, J.; Jin, W.; et al. Na2CO3-responsive mechanisms in halophyte Puccinellia tenuiflora roots revealed by physiological and proteomic analyses. Sci. Rep. 2016, 6, 32717. [Google Scholar] [CrossRef] [PubMed]
- Suo, J.; Zhao, Q.; Zhang, Z.; Chen, S.; Cao, J.; Liu, G.; Wei, X.; Wang, T.; Yang, C.; Dai, S. Cytological and proteomic analyses of Osmunda cinnamomea germinating spores reveal characteristics of fern spore germination and rhizoid tip-growth. Mol. Cell. Proteom. 2015, 14, 2510. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wei, S.; Bian, Y.; Zhao, Q.; Chen, S.; Mao, J.; Song, C.; Cheng, K.; Xiao, Z.; Zhang, C.; Ma, W.; et al. Salinity-induced palmella formation mechanism in halotolerant algae Dunaliella salina revealed by quantitative proteomics and phosphoproteomics. Front. Plant Sci. 2017, 8, 810. [Google Scholar] [CrossRef] [PubMed]
- Matrix Science. Available online: http://www.matrixscience.com (accessed on 2 December 2016).
- National Center for Biotechnology Information Non-Redundant (NCBInr) Protein Databases. Available online: http://www.ncbi.nlm.nih.gov/protein/ (accessed on 2 December 2016).
- UniProt. Available online: http://www.uniprot.org/ (accessed on 15 March 2017).
- YLoc: Interpretable Subcellular Localization Prediction. Available online: http://abi.inf.uni-tuebingen.de/Services/YLoc/webloc.cgi (accessed on 25 March 2017).
- LocTree3: Protein Subcellular Localization Prediction System. Available online: https://rostlab.org/services/loctree3/ (accessed on 25 March 2017).
- Plant-mPLoc: Predicting Subcellular Localization of Plant Proteins Including Those With Multiple Sites. Available online: http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/# (accessed on 25 March 2017).
- ngLOC: A Bayesian method for Predicting Protein Subcellular Localization. Available online: http://genome.unmc.edu/ngLOC/index.html (accessed on 25 March 2017).
- TargetP 1.1 Server. Available online: http://www.cbs.dtu.dk/services/TargetP/ (accessed on 25 March 2017).

| Spot No. (a) | Protein Name (b) | Abbreviation | Subcellular Location (c) | Accession No. (d) | Sco (e) | QM (f) | V%±SD (g) 0 12 24 36 |
|---|---|---|---|---|---|---|---|
| Photosynthetic electron transfer chain (9) | |||||||
| 4044 | Chlorophyll A/B binding protein | CAB | Chl | AAA18529 | 135 | 2 |  |
| 3965 | Chlorophyll A/B binding protein | CAB | Chl | AAA18529 | 153 | 2 |  |
| 4040 | Light harvesting chlorophyll A/B binding protein | CAB | Chl | ABW70800 | 213 | 3 |  |
| 4079 | Chlorophyll A-B binding family protein* | CAB | Chl | ABK96765 | 201 | 3 |  |
| 4064 | Photosystem II oxygen-evolving complex protein 2 precursor | OEC2 | Chl | XP_002300858 | 136 | 2 |  |
| 4192 | Photosynthetic NDH subunit of luminal location 5, chloroplastic* | PnsL5 | Chl | CDP13378 | 102 | 2 |  |
| 3848 | Ferredoxin-NADP reductase, chloroplastic* | FNR | Chl | OAY57547 | 310 | 5 |  |
| 3375 | ATP synthase CF1 α subunit, chloroplastic | cATP synthase α | Chl | AKF33906 | 369 | 6 |  |
| 3104 | ATP synthase CF1 α subunit, chloroplastic | cATP synthase α | Chl | AKF33906 | 164 | 2 |  |
| Calvin cycle (28) | |||||||
| 4038 | Carbonic anhydrase isoform 1* | CA1 | Chl | ABK96336 | 187 | 3 |  |
| 3549 | Ribulose-bisphosphate carboxylase (RuBisCO) activase, chloroplastic | RCA | Chl | Q01587 | 316 | 4 |  |
| 4441 | RuBisCO activase 1, chloroplastic | RCA1 | Chl | Q7X9A0 | 162 | 2 |  |
| 3485 | RuBisCO activase, chloroplastic isoform X1* | RCA X1 | Chl | ABK96359 | 543 | 6 |  |
| 3487 | RuBisCO activase, chloroplastic isoform X1* | RCA X1 | Chl | ABK96359 | 401 | 6 |  |
| 3477 | Rubisco activase isoform 2* | RCA2 | Chl | CDP00127 | 294 | 5 |  |
| 4075 | RuBisCO activase, chloroplastic isoform X2 | RCA X2 | Chl | XP_011002635 | 294 | 5 |  |
| 3777 | RuBisCO large chain | RBCL | Chl | O78258 | 176 | 2 |  |
| 3303 | RuBisCO large subunit | RBCL | Chl | CUR00003 | 176 | 2 |  |
| 3497 | RuBisCO large chain | RBCL | Chl | O78258 | 108 | 2 |  |
| 4259 | RuBisCO small chain | RBCS | Chl | XP_011035062 | 243 | 6 |  |
| 4263 | RuBisCO small chain | RBCS | Chl | XP_002531624 | 112 | 2 |  |
| 3584 | Phosphoglycerate kinase 1 family protein | PGK1 | Chl | XP_002315066 | 351 | 7 |  |
| 3491 | Phosphoglycerate kinase 1 family protein | PGK1 | Chl | XP_002315066 | 351 | 7 |  |
| 3687 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic* | GAPDH-A | Chl | ABK94956 | 484 | 6 |  |
| 3671 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic* | GAPDH-A | Chl | OIW03351 | 166 | 2 |  |
| 3670 | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic* | GAPDH-A | Chl | OIW03351 | 113 | 2 |  |
| 3817 | Fructose-bisphosphate aldolase 1, chloroplastic* | FBA1 | Chl | ABK95613 | 164 | 3 |  |
| 3813 | Fructose-bisphosphate aldolase 3 | FBA3 | Chl | AGB05600 | 185 | 3 |  |
| 3816 | Fructose-bisphosphate aldolase 3 | FBA3 | Chl | AGB05600 | 196 | 3 |  |
| 3809 | Fructose-bisphosphate aldolase 3 | FBA3 | Chl | AGB05600 | 124 | 2 |  |
| 3802 | Fructose-bisphosphate aldolase 3 | FBA3 | Chl | AGB05600 | 133 | 2 |  |
| 3082 | Transketolase, chloroplastic | TK | Chl | KHG01555 | 99 | 2 |  |
| 3114 | Transketolase, chloroplastic | TK | Chl | EMT02862 | 120 | 2 |  |
| 4084 | Transketolase, chloroplastic | TK | Chl | Q43848 | 92 | 2 |  |
| 3639 | Sedoheptulose-1,7-bisphosphatase, chloroplastic* | SBPase | Chl | XP_002316235 | 57 | 2 |  |
| 3580 | Sedoheptulose-1,7-bisphosphatase, chloroplastic* | SBPase | Chl | OAY38482 | 101 | 3 |  |
| 3696 | Phosphoribulokinase, chloroplastic* | PRK | Chl | AOL56425 | 92 | 2 |  |
| Carbohydrate and energy metabolism (12) | |||||||
| 3208 | Phosphoglucomutase, cytoplasmic | PGM | Cyt | Q9ZSQ4 | 115 | 3 |  |
| 3725 | Glyceraldehyde-3-phosphate dehydrogenase 2, cytosolic* | GAPDH2 | Cyt | XP_002318114 | 374 | 4 |  |
| 3256 | Enolase | ― | Cyt | Q42971 | 86 | 2 |  |
| 3506 | Alcohol dehydrogenase* | ADH | Cyt | XP_002302195 | 133 | 5 |  |
| 3500 | Alcohol dehydrogenase* | ADH | Cyt | XP_002302195 | 171 | 3 |  |
| 3701 | NAD-dependent malate dehydrogenase | NAD-MDH | Cyt | AAL11502 | 161 | 2 |  |
| 3702 | NAD-dependent malate dehydrogenase | NAD-MDH | Cyt | AAL11502 | 104 | 2 |  |
| 3710 | NAD-dependent malate dehydrogenase | NAD-MDH | Cyt | AAL11502 | 223 | 3 |  |
| 4448 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial* | PDH E1-β | Mit | GAU16570 | 249 | 3 |  |
| 3409 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial* | PDH E1-β | Mit | GAU16570 | 221 | 3 |  |
| 4202 | ATP synthase subunit d, mitochondrial * | mATP synthase d | Mit | CBI31501 | 89 | 2 |  |
| 4417 | ATP synthase subunit beta, mitochondrial * | mATP synthase β | Mit | CDP00716 | 66 | 2 |  |
| Other metabolism (10) | |||||||
| 3611 | Glutamine synthetase | GS | Cyt | AGG19203 | 179 | 3 |  |
| 3604 | Glutamine synthetase | GS | Cyt | ABF06665 | 150 | 3 |  |
| 3841 | O-acetylserine (thiol) lyase family protein | OAS-TL | Uncertain | XP_006389317 | 70 | 2 |  |
| 3747 | 3-isopropyl malate dehydrogenase, chloroplastic | IPMDH | Chl | P29696 | 117 | 2 |  |
| 3402 | Alanine aminotransferase family protein | ALT | Uncertain | ALT55639 | 239 | 6 |  |
| 3515 | Aldolase superfamily protein isoform 1, delta-aminolevulinic acid dehydratase 1, chloroplastic* | ALAD1 | Chl | EOY16322 | 92 | 3 |  |
| 3587 | Uroporphyrinogen decarboxylase | UROD | Chl | XP_011012304 | 272 | 4 |  |
| 3478 | Anthocyanidin 3-O-glucosyltransferase 2* | A3GT 2 | Uncertain | ABK96136 | 212 | 3 |  |
| 3798 | Isoflavone reductase family protein* | IFR | Cyt | ABK95019 | 133 | 3 |  |
| 3791 | 2-methylene-furan-3-one reductase* | MFOR | Chl | ABK96279 | 238 | 3 |  |
| Protein synthesis (1) | |||||||
| 3071 | Elongation factor G-2, chloroplastic* | EF-G2 | Chl | XP_002304430 | 108 | 4 |  |
| Protein folding and unfolding (8) | |||||||
| 3245 | RuBisCO large subunit-binding protein subunit α | RBP-α | Chl | XP_011000529 | 99 | 3 |  |
| 3133 | Stromal 70 kDa heat shock-related family protein | cpHSP70 | Chl | XP_006389517 | 606 | 7 |  |
| 3102 | Chaperone DnaK | DnaK | Chl | KVI03056 | 445 | 7 |  |
| 3106 | Chaperone DnaK | DnaK | Chl | KVI03056 | 275 | 4 |  |
| 4395 | Heat shock cognate protein 80 | HSC80 | Cyt | P36181 | 87 | 2 |  |
| 4397 | Heat shock protein 90 | HSP90 | Cyt | KVI10442 | 74 | 2 |  |
| 4398 | Heat shock protein 90 | HSP90 | Cyt | KVI10442 | 71 | 2 |  |
| 3814 | Peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic* | CYP26-2 | Chl | XP_002318560 | 281 | 4 |  |
| Redox homeostasis and stress defense (4) | |||||||
| 4086 | Glutathione S-transferase U30 | GST-U30 | Cyt | ANO39995 | 74 | 2 |  |
| 4073 | Glutathione S-transferase F1 | GST-F1 | Cyt | ANO39924 | 164 | 2 |  |
| 3748 | Glyoxalase I homolog family protein | GlyI | Cyt | XP_002305564 | 88 | 2 |  |
| 3636 | Aldo/keto reductase family protein | AKR | Chl | XP_002302125 | 296 | 4 |  |
| Signaling (2) | |||||||
| 4249 | Nucleoside diphosphate kinase 1* | NDPK1 | Cyt | ABK95604 | 139 | 25% |  |
| 3985 | 14-3-3-like protein B* | 14-3-3 B | Cyt | XP_002306545 | 119 | 12% |  |
| Cell structure (2) | |||||||
| 3419 | Tubulin α chain | Tubulin-α | Cyt | Q9FT36 | 101 | 12% |  |
| 4001 | Plastid-lipid-associated protein 6, chloroplastic* | PAP6 | Chl | AAR26489 | 154 | 18% |  |
| Miscellaneous or function unknown (5) | |||||||
| 3898 | Haloacid dehalogenase-like hydrolase family protein* | HAD | Chl | ABK96272 | 114 | 7% |  |
| 4457 | Pop3 peptide family protein* | Pop3 | Uncertain | 1SI9_A | 207 | 51% |  |
| 3662 | Soybean genes regulated by cold 2-like domain* | SRC2 | Uncertain | XP_011030162 | 136 | 10% |  |
| 4278 | Stress responsive A/B barrel domain* | SRB | Chl | CAA39082 | 132 | 24% |  |
| 3615 | Unknown protein | ― | Secreted | ABK94923 | 93 | 7% |  |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Jin, X.; Sun, X.; Gao, T.; Chen, X.; She, Y.; Jiang, T.; Chen, S.; Dai, S. Hydrogen Peroxide Response in Leaves of Poplar (Populus simonii × Populus nigra) Revealed from Physiological and Proteomic Analyses. Int. J. Mol. Sci. 2017, 18, 2085. https://doi.org/10.3390/ijms18102085
Yu J, Jin X, Sun X, Gao T, Chen X, She Y, Jiang T, Chen S, Dai S. Hydrogen Peroxide Response in Leaves of Poplar (Populus simonii × Populus nigra) Revealed from Physiological and Proteomic Analyses. International Journal of Molecular Sciences. 2017; 18(10):2085. https://doi.org/10.3390/ijms18102085
Chicago/Turabian StyleYu, Juanjuan, Xin Jin, Xiaomei Sun, Tianxiang Gao, Xiaomei Chen, Yimin She, Tingbo Jiang, Sixue Chen, and Shaojun Dai. 2017. "Hydrogen Peroxide Response in Leaves of Poplar (Populus simonii × Populus nigra) Revealed from Physiological and Proteomic Analyses" International Journal of Molecular Sciences 18, no. 10: 2085. https://doi.org/10.3390/ijms18102085