Overexpression of Populus trichocarpa Mitogen-Activated Protein Kinase Kinase4 Enhances Salt Tolerance in Tobacco
Abstract
:1. Introduction
2. Result
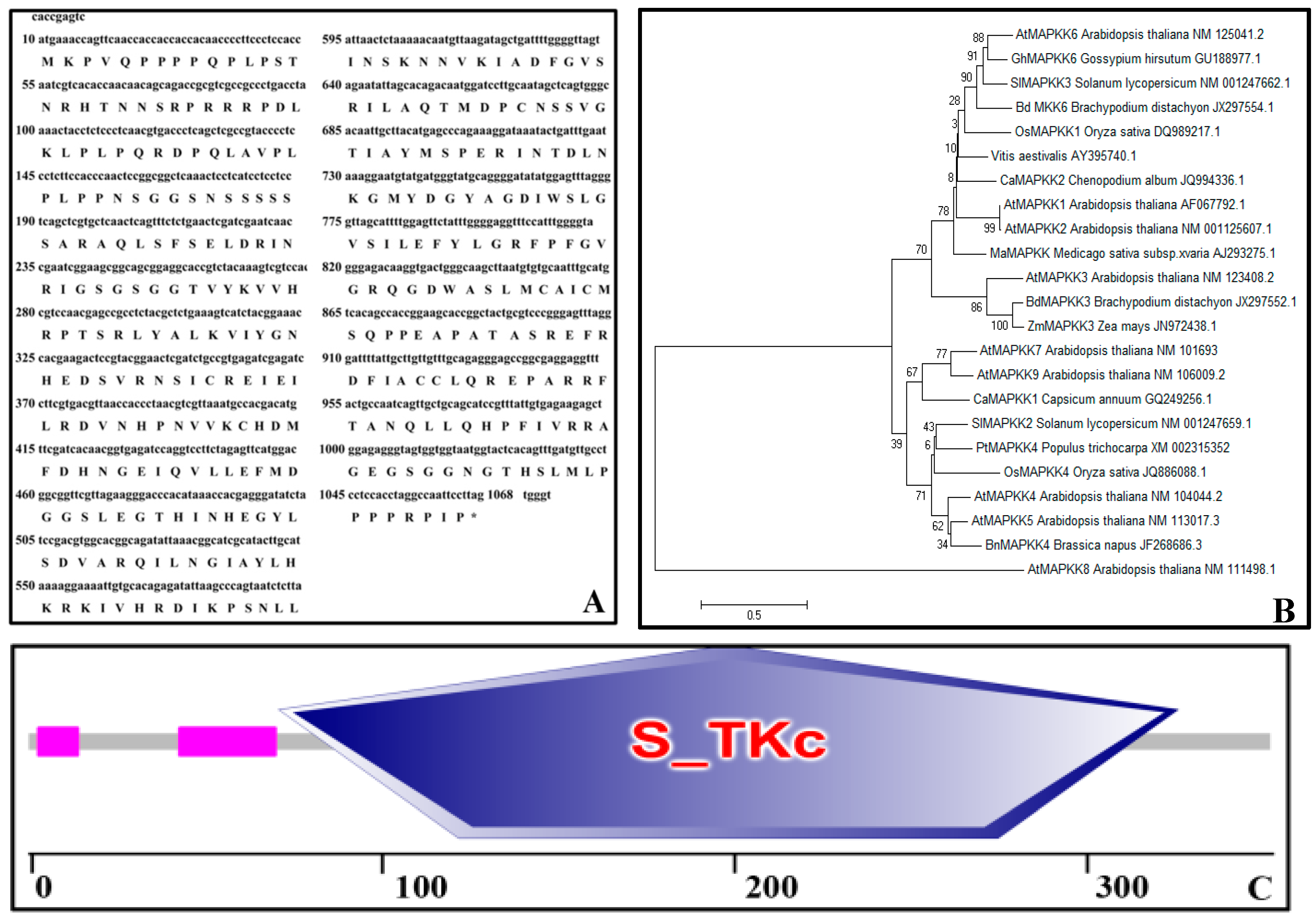
2.1. Bioinformatics Analysis of PtMAPKK4 Sequence
2.2. PtMAPKK4 Gene Expression Properties
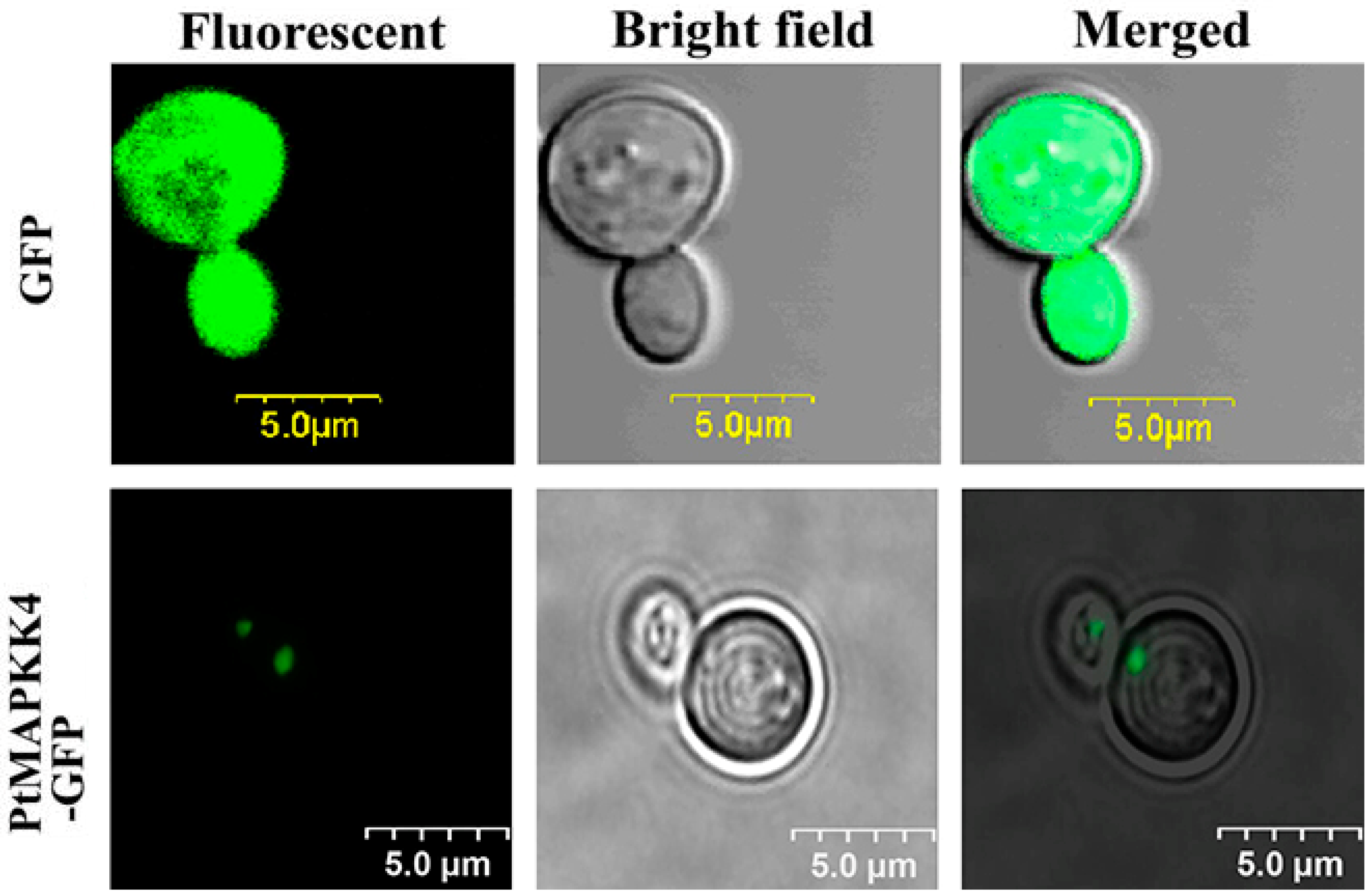
2.3. Subcellular Localization of PtMAKK4 Protein
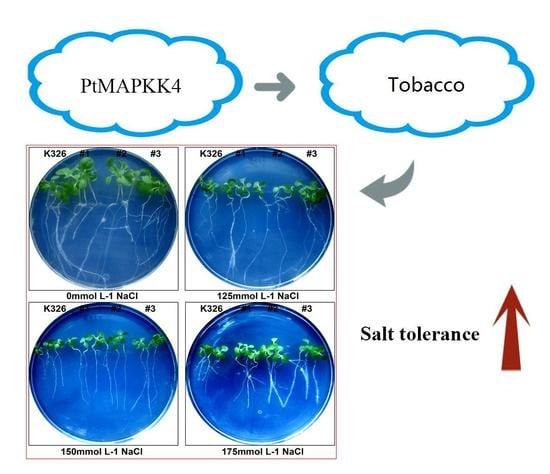
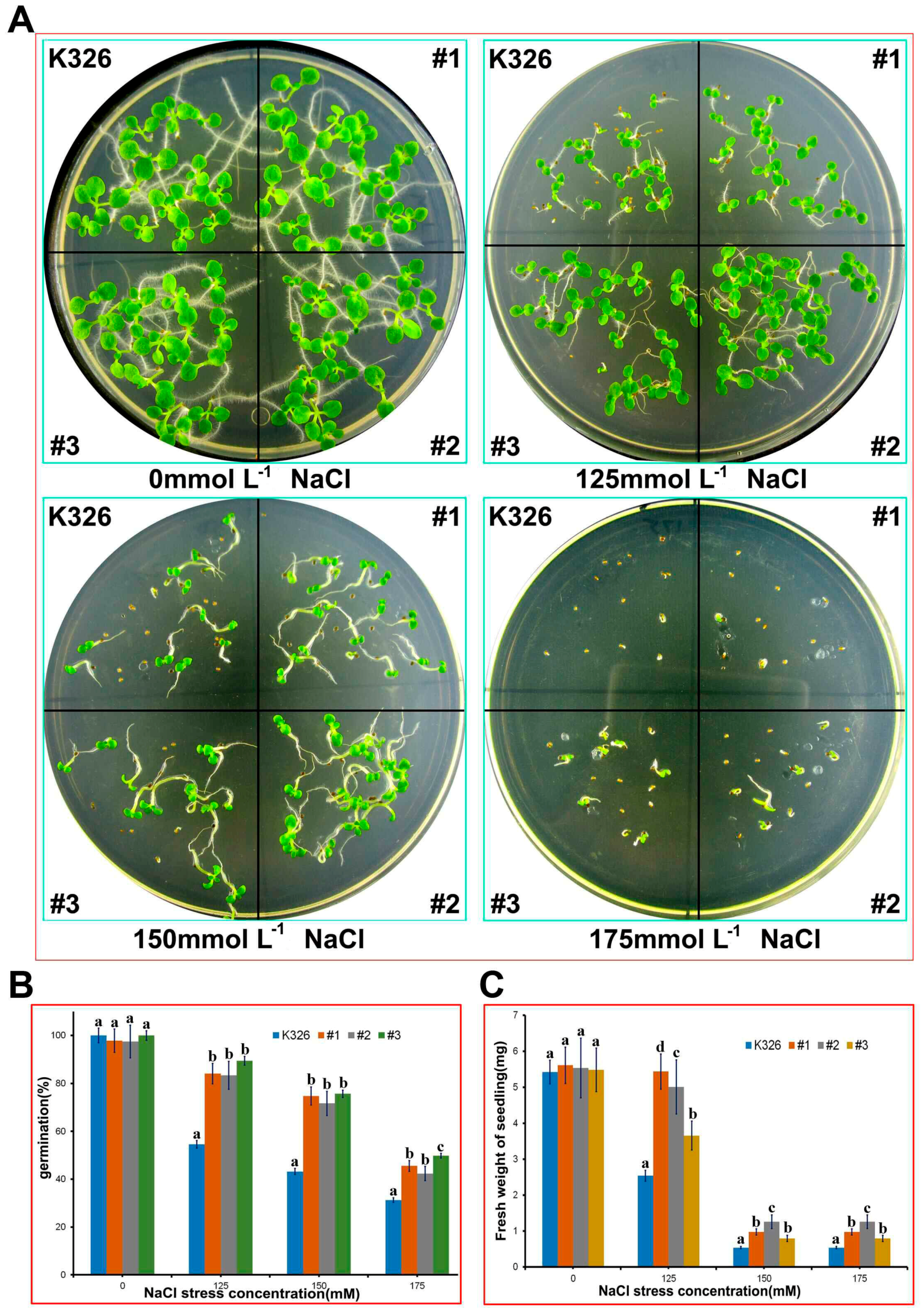
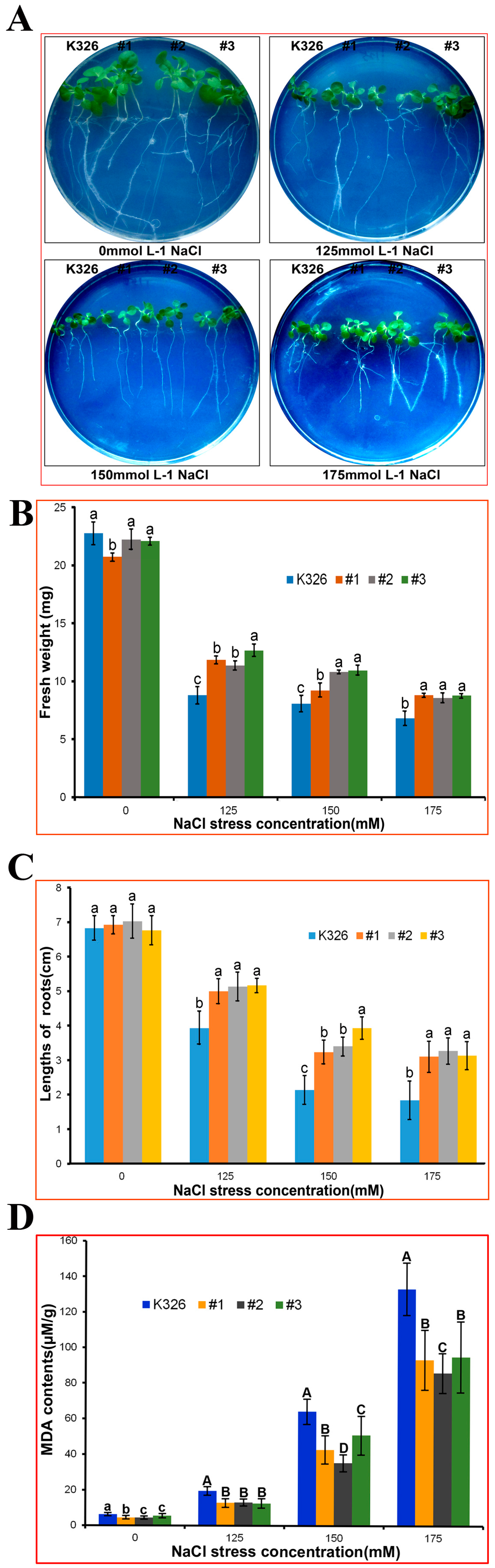
2.4. Analysis of PtMAPKK4 Gene Overexpression in Response to Salt Tolerance
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction and Preparation of cDNA
4.3. Gene Cloning and Plasmid Constructions
4.4. Bioinformation Analysis
4.5. qRT-PCR Analysis
4.6. Subcellular Localization Analysis
4.7. RNA Gel Blot Assay
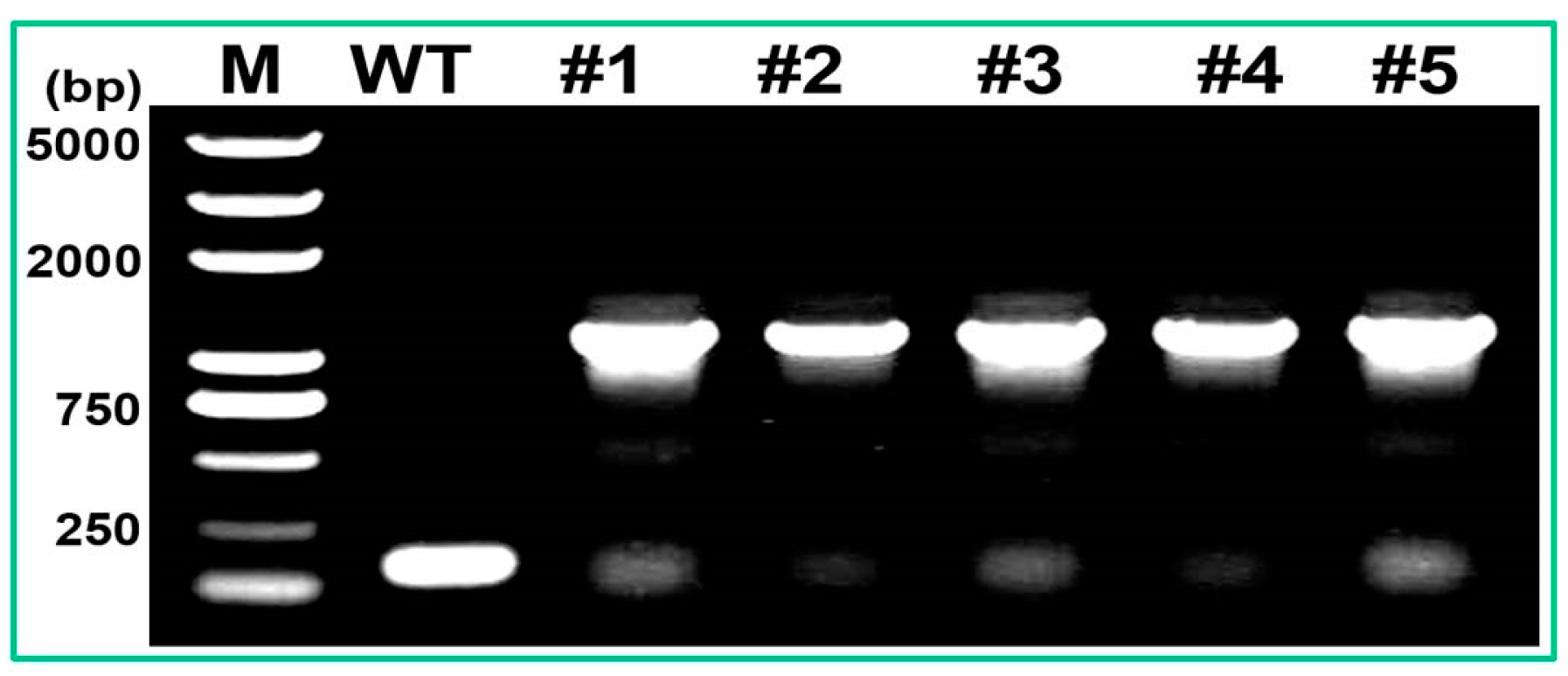
4.8. Plant Genetic Transformation and Analysis of Salt Tolerance
4.9. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ning, J.; Li, X.; Hicks, L.M.; Xiong, L. A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 2010, 152, 876–890. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Abscisic Acid and Abiotic Stress Signaling. Plant Signal. Behav. 2007, 2, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Okrész, L.; Bögre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin.Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Widmann, C.; Gibson, S.; Jarpe, M.B.; Johnson, G.L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 1999, 79, 143–180. [Google Scholar] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomezgomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Pitzschke, A.; Hirt, H. Emerging MAP kinase pathways in plant stress signalling. Trend. Plant Sci. 2005, 10, 339. [Google Scholar] [CrossRef] [PubMed]
- Nicole, M.C.; Hamel, L.P.; Morency, M.J.; Beaudoin, N.; Ellis, B.E.; Séguin, A. MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genom. 2006, 7, 223. [Google Scholar] [CrossRef] [PubMed]
- Kazuya, L.; Kazuo, S.; Guillaume, T.; Jen, S.; Yves, H.; Anthony, C.; Martin, K.; Shuqun, Z.; Heribert, H.; Cathal, W.; et al. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trend. Plant Sci. 2002, 7, 301. [Google Scholar]
- Burnett, E.C.; Desikan, R.; Moser, R.C.; Neill, S.J. ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 2000, 51, 197–205. [Google Scholar] [CrossRef]
- Knetsch, M.; Wang, M.; Snaarjagalska, B.E.; Heimovaaradijkstra, S. Abscisic Acid Induces Mitogen-Activated Protein Kinase Activation in Barley Aleurone Protoplasts. Plant Cell 1996, 8, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Mockaitis, K.; Howell, S.H. Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 2000, 24, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Nishihama, R.; Morikiyo, K.; Ishikawa, M.; Machida, Y. NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 2003, 17, 1055. [Google Scholar] [CrossRef] [PubMed]
- Voronin, V.; Aionesei, T.; Limmongkon, A.; Barinova, I.; Touraev, A.; Laurière, C.; Coronado, M.J.; Testillano, P.S.; Risueño, M.C.; Heberle-Bors, E. The MAP kinase kinase NtMEK2 is involved in tobacco pollen germination. FEBS Lett. 2004, 560, 86–90. [Google Scholar] [CrossRef]
- Jonak, C.; Nakagami, H.; Hirt, H. Heavy metal stress. Activation of distinct mitogen-activated protein kinase pathways by copper and cadmium. Plant Physiol. 2004, 136, 3276. [Google Scholar] [CrossRef] [PubMed]
- Mayrose, M.; Bonshtien, A.; Sessa, G. LeMPK3 is a mitogen-activated protein kinase with dual specificity induced during tomato defense and wounding responses. J. Biol. Chem. 2004, 279, 14819. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.A.; Miles, G.P.; Ellis, B.E. Ozone treatment rapidly activates MAP kinase signalling in plants. Plant J. 2000, 22, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Klessig, D.F. MAPK cascades in plant defense signaling. Trend. Plant Sci. 2001, 6, 520. [Google Scholar] [CrossRef]
- Hamel, L.; Nicole, M.; Sritubtim, S.; Morency, M.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D.; Lee, J.; et al. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trend. Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Su, H.; Han, L.; Wang, C.; Sun, Y.; Liu, F. Differential expression profiles of poplar MAP kinase kinases in response to abiotic stresses and plant hormones, and overexpression of PtMKK4 improves the drought tolerance of poplar. Gene 2014, 545, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Rao, K.P.; Sharma, P.; Sinha, A.K. Differential regulation of rice mitogen activated protein kinase kinase (MKK) by abiotic stress. Plant Physiol. Biochem. Ppb 2008, 46, 891. [Google Scholar] [CrossRef] [PubMed]
- Nakagami, H.; Soukupová, H.; Schikora, A.; Zárský, V.; Hirt, H. A Mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 2006, 281, 38697. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Soyano, T.; Kosetsu, K.; Sasabe, M.; Machida, Y. HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1766–1776. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Li, Y.; Lu, W.; Fei, M.; Wu, C.; Guo, X. Cotton GhMKK5 affects disease resistance, induces HR-like cell death, and reduces the tolerance to salt and drought stress in transgenic Nicotiana benthamiana. J. Exp. Bot. 2012, 63, 3935. [Google Scholar]
- Nishihama, R.; Banno, H.; Shibata, W.; Hirano, K.; Nakashima, M.; Usami, S.; Machida, Y. Plant Homologues of Components of MAPK (Mitogen-Activated Protein Kinase) Signal Pathways in Yeast and Animal Cells. Plant Cell Physiol. 1995, 36, 749. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.C.S.; Petersen, M.; Mundy, J. Mitogen-Activated Protein Kinase Signaling in Plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [PubMed]
- Song, Z.; Xuefeng, X.; Jincheng, S.; Zhirong, S. Bioinformatics Research in Subcellular Localization of Protein. Progress Biochem. Biophys. 2007, 34, 573–579. [Google Scholar]
- Song, Z.; Xuefeng, X.; Jincheng, S.; Zhirong, S. Eukaryotic Protein Subcellular Localization Prediction Based on Sequence Conservation and Protein-Protein Interaction. Progress Biochem. Biophys. 2008, 35, 531–535. [Google Scholar]
- Devos, D.; Valencia, A. Practical limits of function prediction. Proteins 2000, 41, 98. [Google Scholar] [CrossRef]
- Ambron, R.T.; Schmied, R.; Huang, C.C.; Smedman, M. A signal sequence mediates the retrograde transport of proteins from the axon periphery to the cell body and then into the nucleus. J. Neurosci. 1992, 12, 2813–2818. [Google Scholar] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 Pathway Mediates Cold and Salt Stress Signaling in Arabidopsis. Mol. Cell 2004, 15, 141. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Xiang, C.B. The genetic locus At1g73660 encodes a putative MAPKKK and negatively regulates salt tolerance in Arabidopsis. Plant Mol. Biol. 2008, 67, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.N.; Qu, Z.C.; Wan, Y.Z.; Zhang, H.W.; Shen, D.L. Application of Suppression Subtractive Hybridization (SSH) to Cloning Differentially Expressed cDNA in Dunaliella salina (Chlorophyta) Under Hyperosmotic Shock. Plant Mol. Biol. Rep. 2002, 20, 49–57. [Google Scholar] [CrossRef]
- Alkayal, F.; Albion, R.L.; Tillett, R.L.; Hathwaik, L.T.; Lemos, M.S.; Cushman, J.C. Expressed sequence tag (EST) profiling in hyper saline shocked Dunaliella salina reveals high expression of protein synthetic apparatus components. Plant Sci. 2010, 179, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Wang, J.; Wang, D.; Fan, W.; Wang, S.; Ye, W. The MAPKKK gene family in Gossypium raimondii: genome-wide identification, classification and expression analysis. Int. J. Mol. Sci. 2013, 14, 18740–18757. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Habor Press Cold Spring Harbor: New York, NY, USA, 1989. [Google Scholar]



| Primer | Primer Sequence |
|---|---|
| Actin2-F | 5′-TTCTACAAGTGCTTTGATGGTGAGTTC-3′ |
| Actin2-R | 5′- CTATTCGATACATAGAAGATCAGAATGTTC-3′ |
| PtMAPKK4-Q-F | 5′-GCCATCGCCATAGTAGTTCTCCT-3′ |
| PtMAPKK4-Q-R | 5′-CATAACCCAAGGCTGTGGAACT-3′ |
| PtMAPKK4-1F | 5′-CACCGAGTCATGAAACCAGTTC-3′ |
| PtMAPKK4-1R | 5′-ACCCACTAAGGAATTGGCCTAG-3′ |
| pYES2-PutMAPK4-G-F | 5′-GGTACCATGAAACCAGTTCAACC-3′ |
| pYES2-PutMAPK4-G-R | 5′-ACTAGTAGGAATTGGCCTAG-3′ |
| pCXSN-PutMAPK4-F | 5′-CACCGAGTCATGAAACCAGTTC-3′ |
| pCXSN-PutMAPK4-R | 5′-ACCCACTAAGGAATTGGCCTAG-3′ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, C.; Wang, R.; Gou, L.; Si, Y.; Guan, Q. Overexpression of Populus trichocarpa Mitogen-Activated Protein Kinase Kinase4 Enhances Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2017, 18, 2090. https://doi.org/10.3390/ijms18102090
Yang C, Wang R, Gou L, Si Y, Guan Q. Overexpression of Populus trichocarpa Mitogen-Activated Protein Kinase Kinase4 Enhances Salt Tolerance in Tobacco. International Journal of Molecular Sciences. 2017; 18(10):2090. https://doi.org/10.3390/ijms18102090
Chicago/Turabian StyleYang, Chengjun, Ruoning Wang, Luzheng Gou, Yongchao Si, and Qingjie Guan. 2017. "Overexpression of Populus trichocarpa Mitogen-Activated Protein Kinase Kinase4 Enhances Salt Tolerance in Tobacco" International Journal of Molecular Sciences 18, no. 10: 2090. https://doi.org/10.3390/ijms18102090