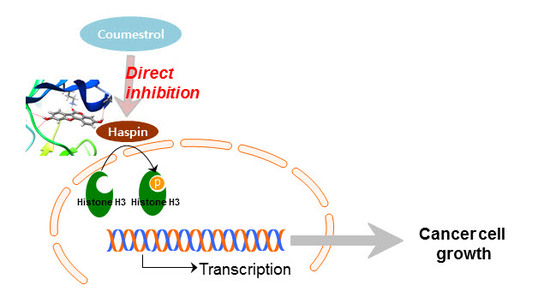
Coumestrol Epigenetically Suppresses Cancer Cell Proliferation: Coumestrol Is a Natural Haspin Kinase Inhibitor
Abstract
:1. Introduction
2. Results
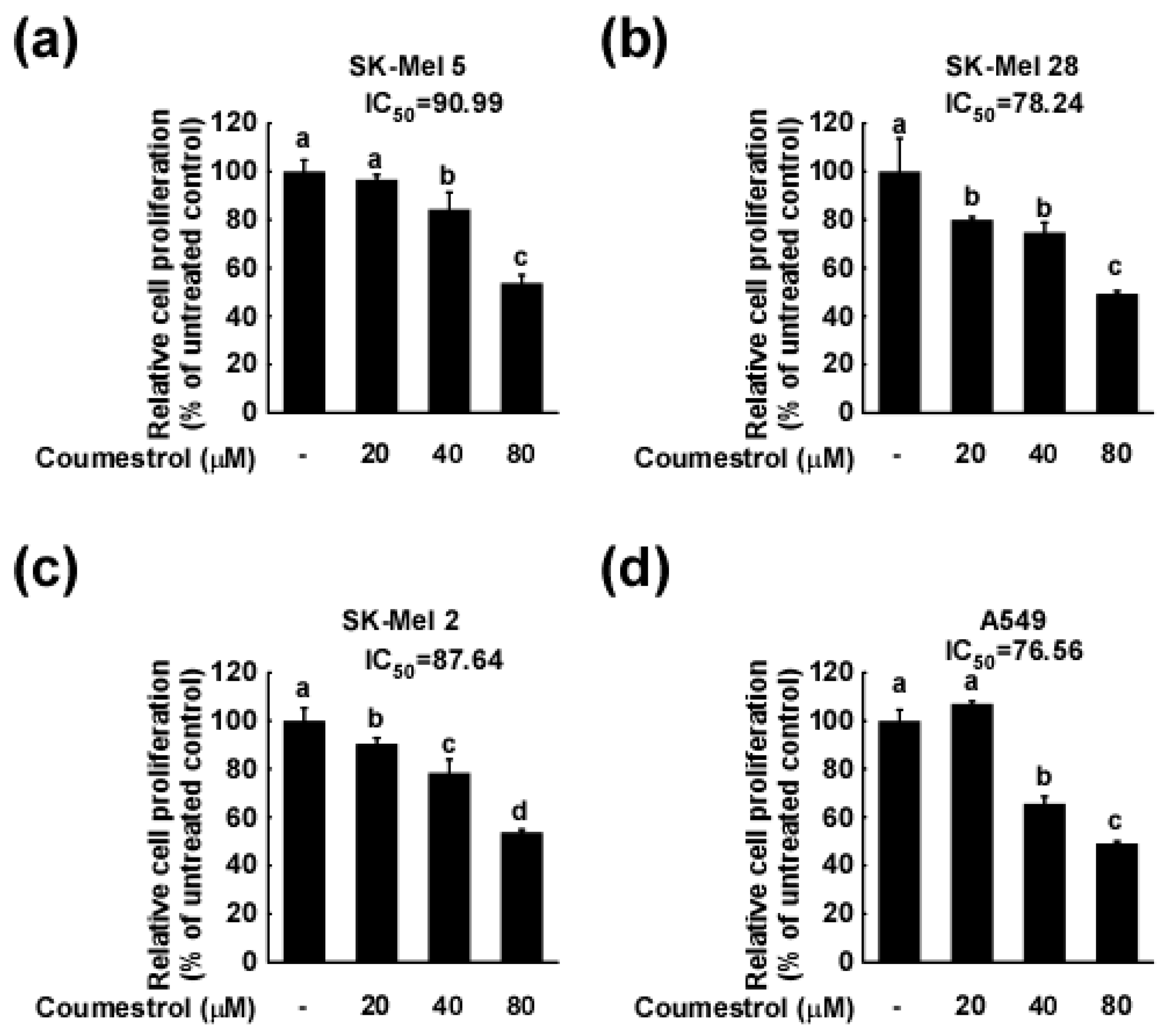
2.1. Coumestrol Inhibits the Growth of Various Cancer Cell Types
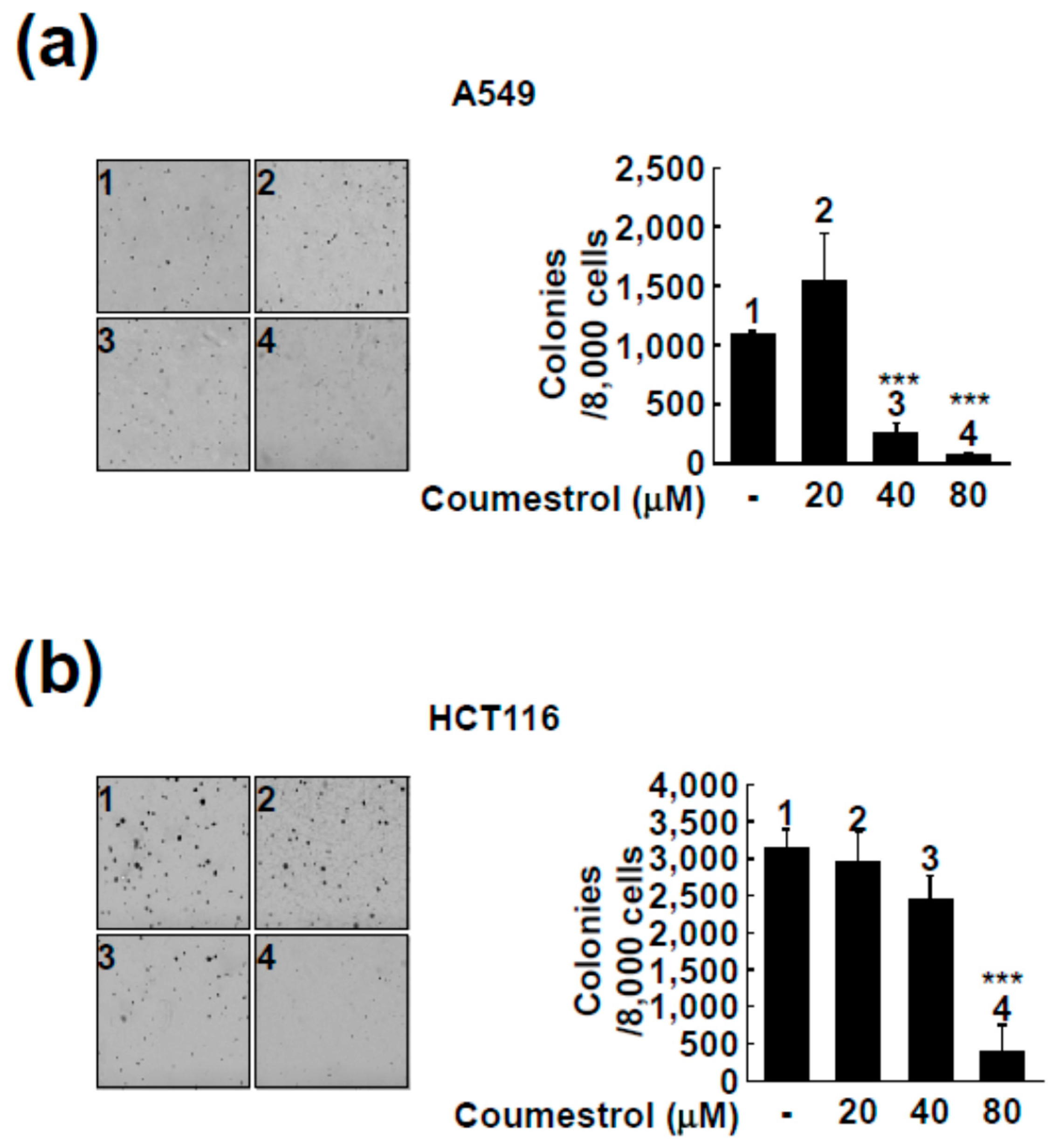
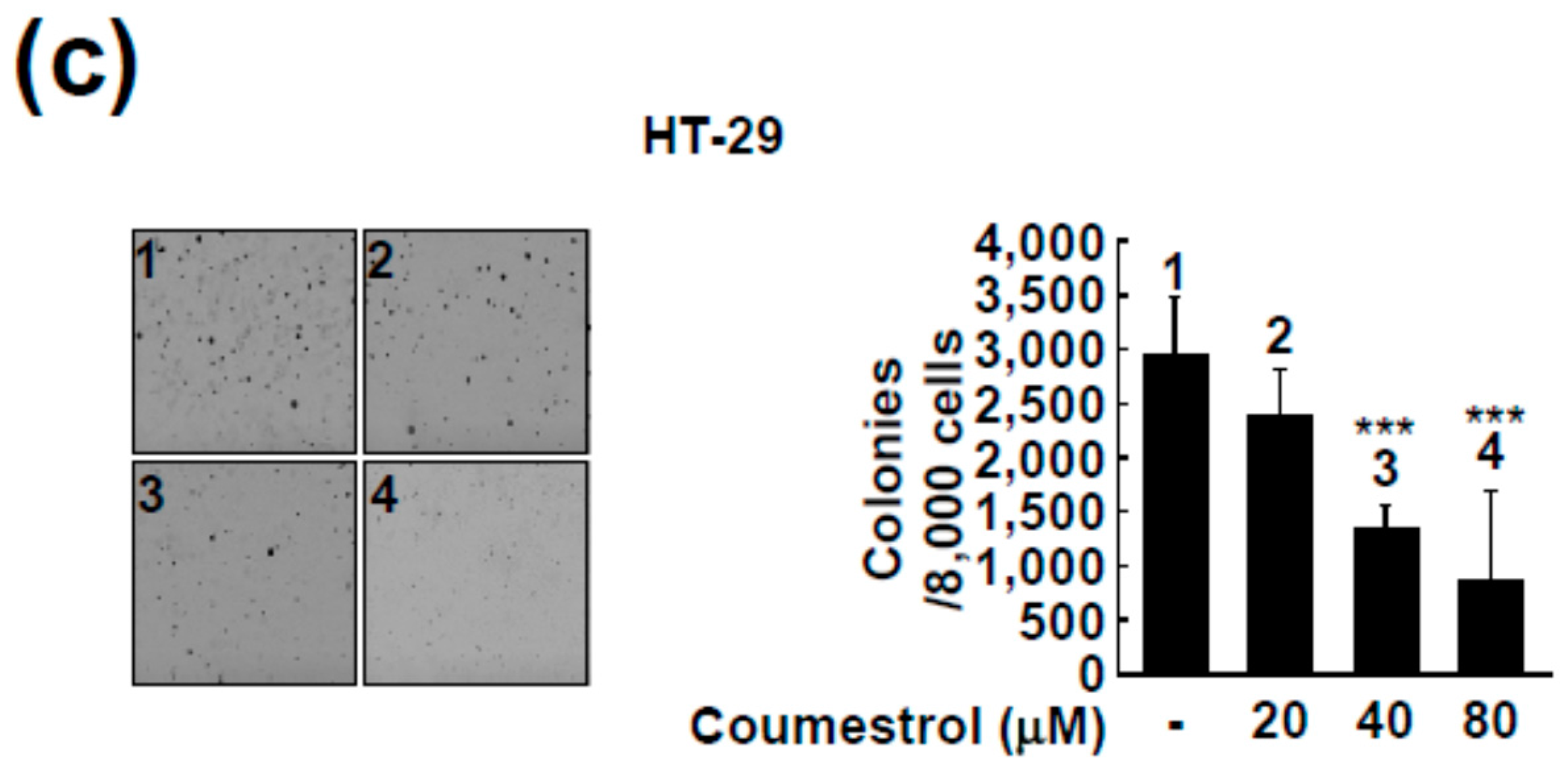
2.2. Coumestrol Reduces Anchorage-Independent Cancer Cell Growth
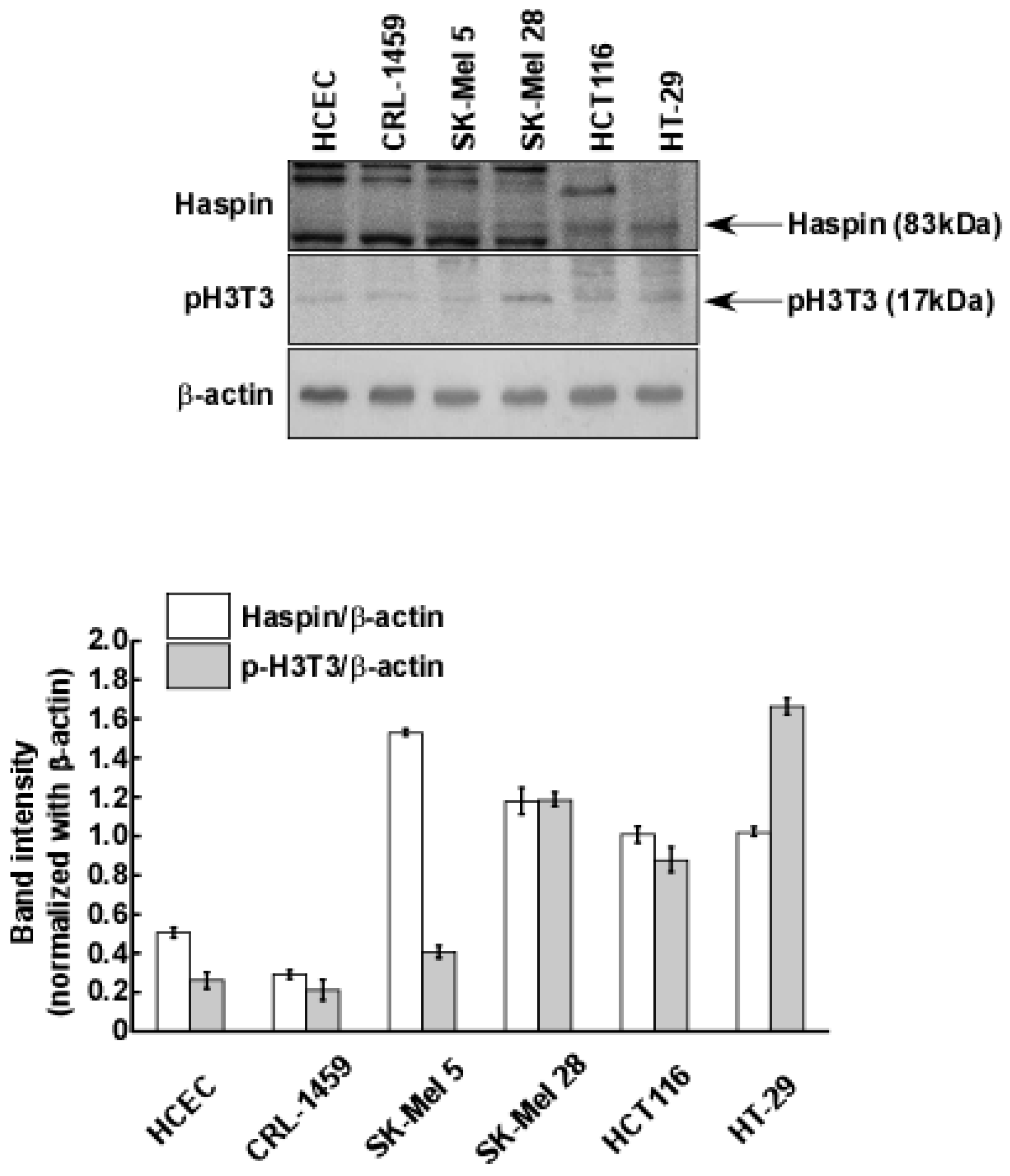
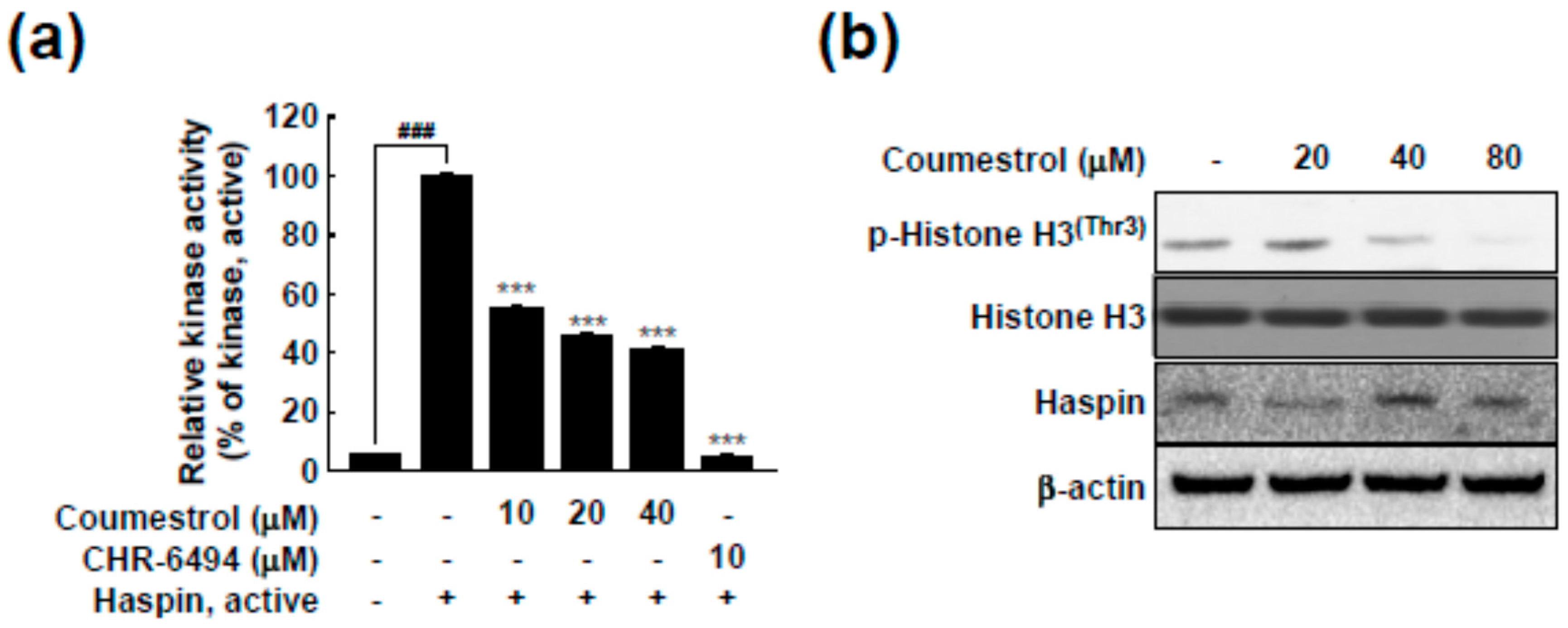
2.3. Haspin Kinase Is a Direct Target of Coumestrol
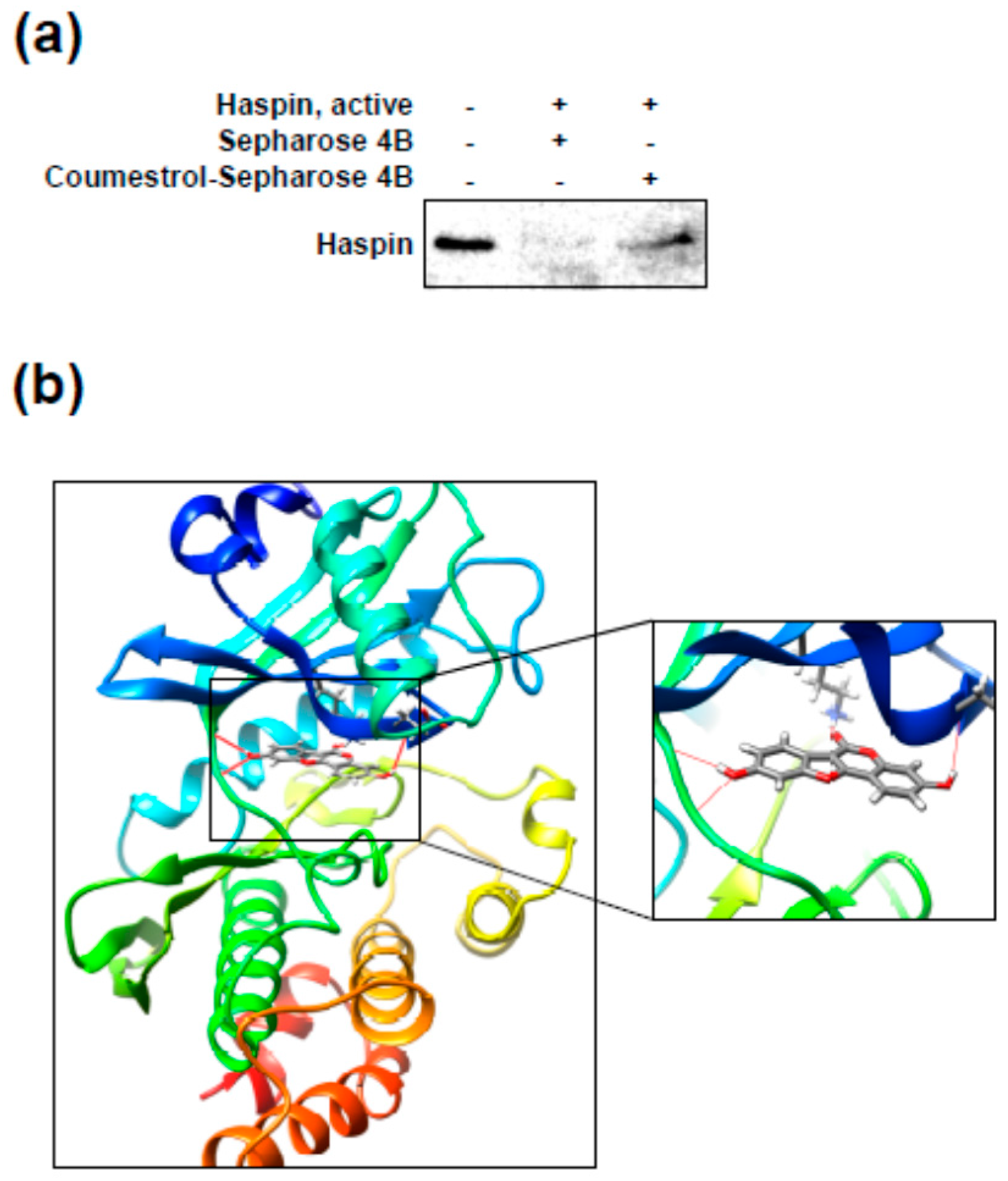
2.4. Coumestrol Directly Interacts with Haspin Kinase
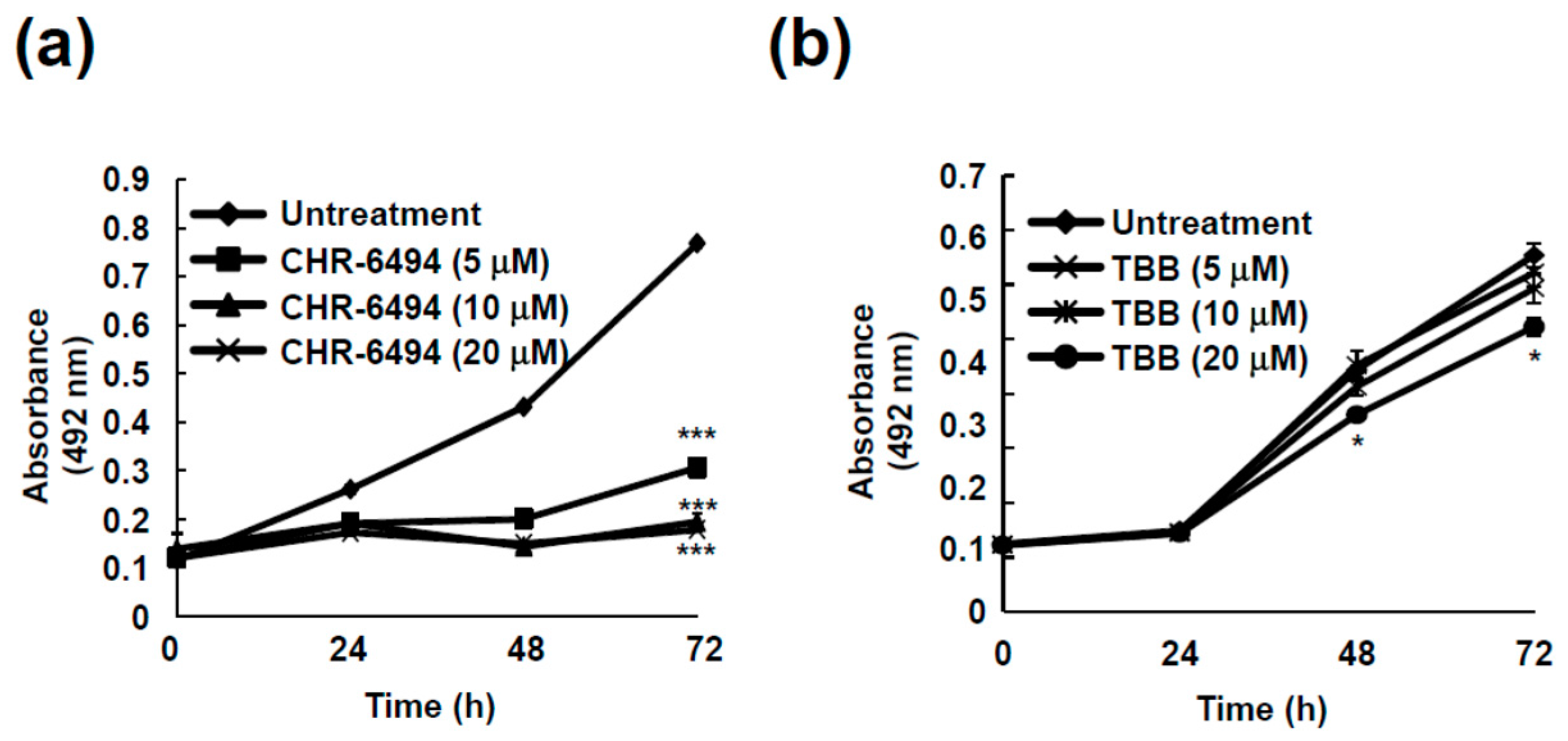
2.5. Haspin Kinase Modulates Cancer Cell Growth but Not Casein Kinase 2
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Cell Proliferation Assay
4.4. Anchorage-Independent Cell Growth
4.5. Preparation of Coumestrol-Sepharose 4B Beads
4.6. Pull down Assay
4.7. Kinase Assay
4.8. Western Blot Analysis
4.9. Molecular Modeling
4.10. Statistical Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stewart, B.W.; Wild, C.P. World Cancer Report 2014; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2015. Ca-Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Oncogenes and cancer. N. Engl. J. Med. 2008, 358, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Schnekenburger, M.; Diederich, M. Epigenetics offer new horizons for colorectal cancer prevention. Curr. Color. Cancer Rep. 2012, 8, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.M. Haspin: A newly discovered regulator of mitotic chromosome behavior. Chromosoma 2010, 119, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.M. The Haspin gene: Location in an intron of the integrin αE gene, associated transcription of an integrin αE-derived RNA and expression in diploid as well as haploid cells. Gene 2001, 267, 55–69. [Google Scholar] [CrossRef]
- Cuny, G.D.; Ulyanova, N.P.; Patnaik, D.; Liu, J.F.; Lin, X.; Auerbach, K.; Ray, S.S.; Xian, J.; Glicksman, M.A.; Stein, R.L.; et al. Structure-activity relationship study of beta-carboline derivatives as haspin kinase inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 2015–2019. [Google Scholar] [CrossRef] [PubMed]
- Cuny, G.D.; Robin, M.; Ulyanova, N.P.; Patnaik, D.; Pique, V.; Casano, G.; Liu, J.F.; Lin, X.; Xian, J.; Glicksman, M.A.; et al. Structure-activity relationship study of acridine analogs as haspin and DYRK2 kinase inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 3491–3494. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Kateneva, A.V.; Higgins, J.M. Studies of haspin-depleted cells reveal that spindle-pole integrity in mitosis requires chromosome cohesion. J. Cell Sci. 2009, 122 Pt 22, 4168–4176. [Google Scholar] [CrossRef]
- Dai, J.; Sullivan, B.A.; Higgins, J.M. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell 2006, 11, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Huertas, D.; Soler, M.; Moreto, J.; Villanueva, A.; Martinez, A.; Vidal, A.; Charlton, M.; Moffat, D.; Patel, S.; McDermott, J.; et al. Antitumor activity of a small-molecule inhibitor of the histone kinase Haspin. Oncogene 2012, 31, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Canal Castro, C.; Pagnussat, A.S.; Orlandi, L.; Worm, P.; Moura, N.; Etgen, A.M.; Alexandre Netto, C. Coumestrol has neuroprotective effects before and after global cerebral ischemia in female rats. Brain Res. 2012, 1474, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Simons, R.; Vincken, J.P.; Bohin, M.C.; Kuijpers, T.F.; Verbruggen, M.A.; Gruppen, H. Identification of prenylated pterocarpans and other isoflavonoids in Rhizopus spp. elicited soya bean seedlings by electrospray ionisation mass spectrometry. RCM 2011, 25, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Ndebele, K.; Graham, B.; Tchounwou, P.B. Estrogenic activity of coumestrol, DDT, and TCDD in human cervical cancer cells. Int. J. Environ. Res. Public Health 2010, 7, 2045–2056. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Yuk, H.J.; Park, K.H.; Bae, Y.S. Coumestrol induces senescence through protein kinase CKII inhibition-mediated reactive oxygen species production in human breast cancer and colon cancer cells. Food Chem. 2013, 141, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.Y.; Cho, S.; Park, E.; Kim, B.; Sohn, E.J.; Oh, B.; Lee, E.O.; Lee, H.J.; Kim, S.H. Coumestrol suppresses hypoxia inducible factor 1α by inhibiting ROS mediated sphingosine kinase 1 in hypoxic PC-3 prostate cancer cells. Bioorg. Med. Chem. Lett. 2014, 24, 2560–2564. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hsieh, D.; Yang, Y.L.; Xu, Z.; Peto, C.; Jablons, D.M.; You, L. Coumestrol from the national cancer Institute’s natural product library is a novel inhibitor of protein kinase CK2. BMC Pharmacol. Toxicol. 2013, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Baek, S.; Kim, J.E.; Lim, T.G.; Lee, C.C.; Yang, H.; Kang, Y.G.; Park, J.S.; Augustin, M.; Mrosek, M.; et al. Flt3 is a target of coumestrol in protecting against UVB-induced skin photoaging. Biochem. Pharmacol. 2015, 98, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Iloki Assanga, S.B.; Gil-Salido, A.A.; Lewis Luján, L.M.; Rosas-Durazo, A.; Acosta-Silva, A.L.; Rivera-Castañeda, E.G.; Rubio-Pino, J.L. Cell growth curves for different cell lines and their relationship with biological activities. Int. J. Biotechnol. Mol. Biol. Res. 2013, 4, 11. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lim, T.G.; Chen, H.; Jung, S.K.; Lee, H.J.; Lee, M.H.; Kim, D.J.; Shin, A.; Lee, K.W.; Bode, A.M.; et al. Esculetin suppresses proliferation of human colon cancer cells by directly targeting beta-catenin. Cancer Prev. Res. (Phila.) 2013, 6, 1356–1364. [Google Scholar] [CrossRef] [PubMed]
- Lim, T.G.; Lee, S.Y.; Huang, Z.; Lim do, Y.; Chen, H.; Jung, S.K.; Bode, A.M.; Lee, K.W.; Dong, Z. Curcumin suppresses proliferation of colon cancer cells by targeting CDK2. Cancer Prev. Res. (Phila.) 2014, 7, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurihara, D.; Matsunaga, S.; Omura, T.; Higashiyama, T.; Fukui, K. Identification and characterization of plant Haspin kinase as a histone H3 threonine kinase. BMC Plant Biol. 2011, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Schrödinger. Schrödinger Suite 2015; Schrödinger, LLC: New York, NY, USA, 2015. [Google Scholar]
- Eswaran, J.; Patnaik, D.; Filippakopoulos, P.; Wang, F.W.; Stein, R.L.; Murray, J.W.; Higgins, J.M.G.; Knapp, S. Structure and functional characterization of the atypical human kinase haspin. Proc. Natl. Acad. Sci. USA 2009, 106, 20198–20203. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-E.; Lee, S.-Y.; Jang, M.; Choi, H.-K.; Kim, J.H.; Chen, H.; Lim, T.-G.; Dong, Z.; Lee, K.W. Coumestrol Epigenetically Suppresses Cancer Cell Proliferation: Coumestrol Is a Natural Haspin Kinase Inhibitor. Int. J. Mol. Sci. 2017, 18, 2228. https://doi.org/10.3390/ijms18102228
Kim J-E, Lee S-Y, Jang M, Choi H-K, Kim JH, Chen H, Lim T-G, Dong Z, Lee KW. Coumestrol Epigenetically Suppresses Cancer Cell Proliferation: Coumestrol Is a Natural Haspin Kinase Inhibitor. International Journal of Molecular Sciences. 2017; 18(10):2228. https://doi.org/10.3390/ijms18102228
Chicago/Turabian StyleKim, Jong-Eun, Sung-Young Lee, Mi Jang, Hyo-Kyung Choi, Jong Hun Kim, Hanyong Chen, Tae-Gyu Lim, Zigang Dong, and Ki Won Lee. 2017. "Coumestrol Epigenetically Suppresses Cancer Cell Proliferation: Coumestrol Is a Natural Haspin Kinase Inhibitor" International Journal of Molecular Sciences 18, no. 10: 2228. https://doi.org/10.3390/ijms18102228