Phospholamban Is Downregulated by pVHL-Mediated Degradation through Oxidative Stress in Failing Heart
Abstract
:1. Introduction
2. Results
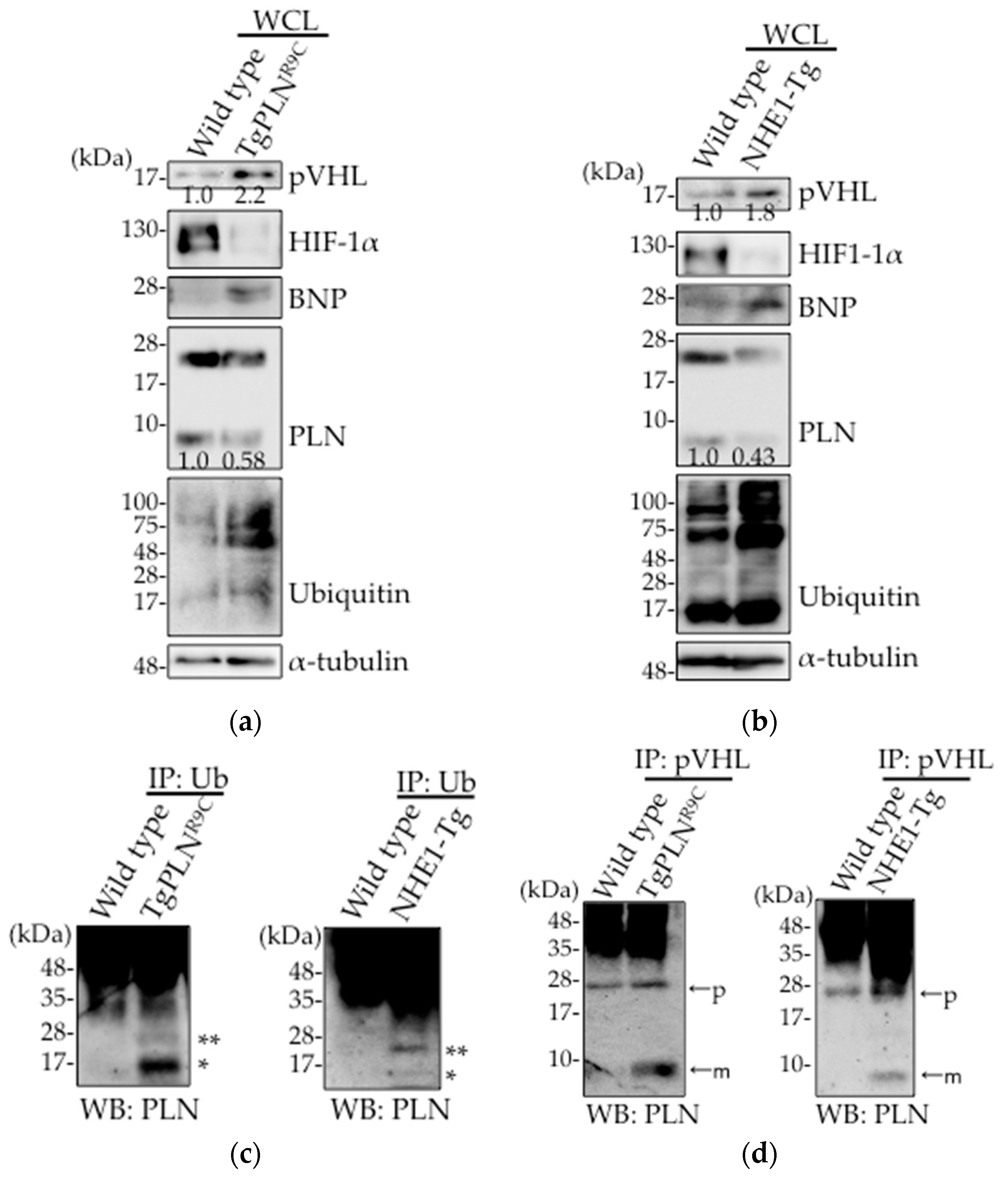
2.1. Upregulation of pVHL Expression and Its Interaction with PLN in Heart Tissues in Two Types of DCM Mice Models, TgPLNR9C and NHE1-Tg Mice
2.2. pVHL Contributes to the Ubiquitination of PLN
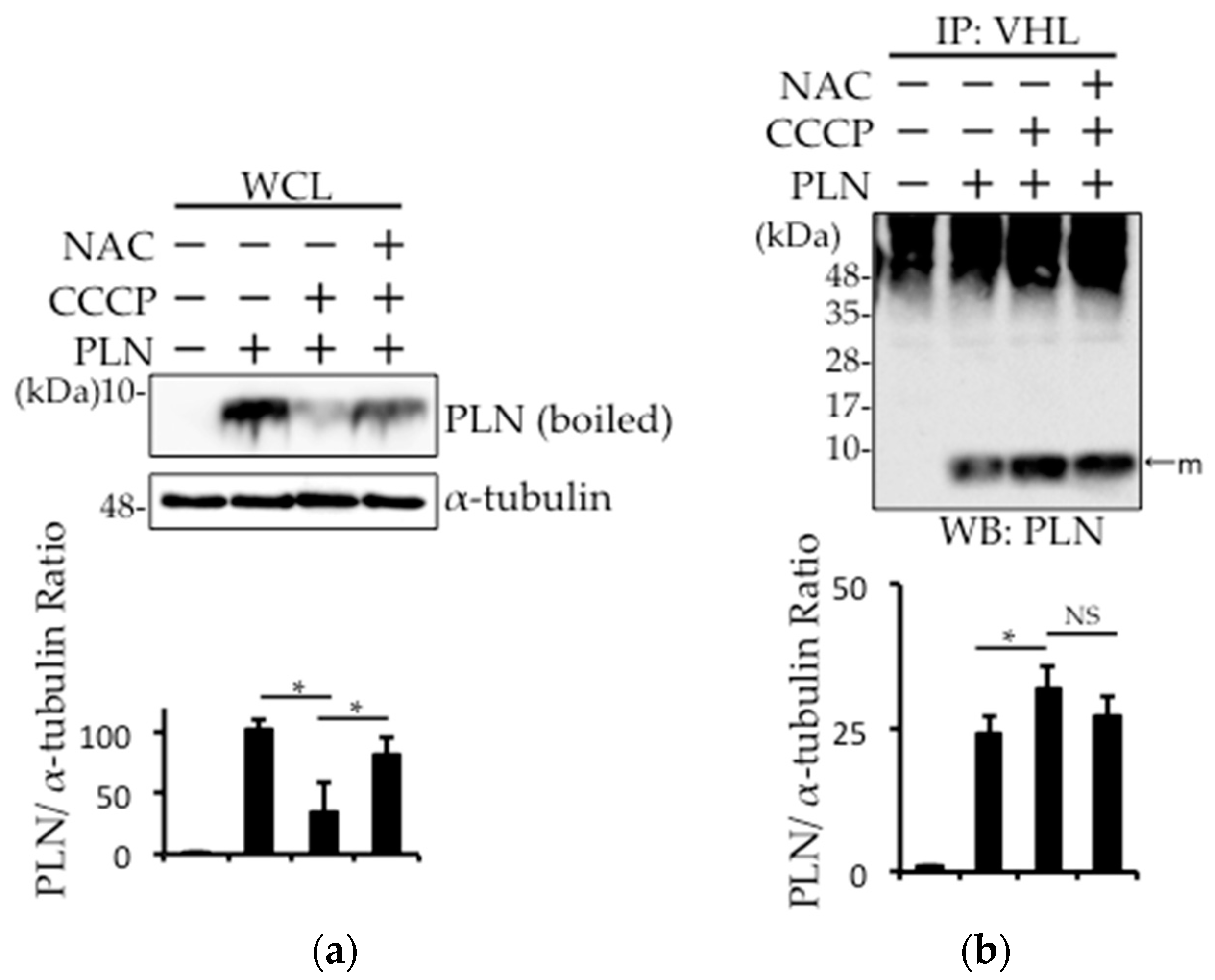
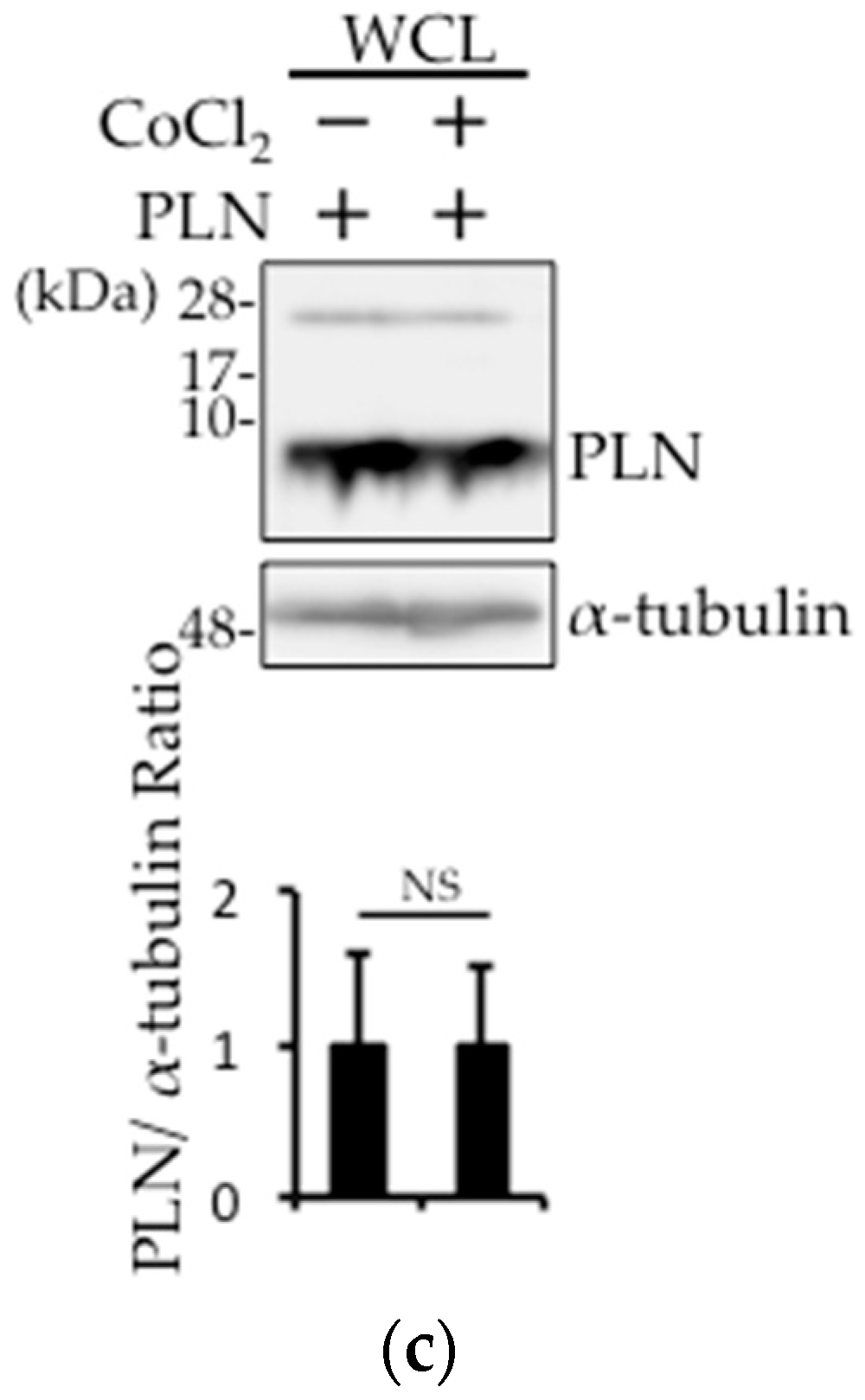
2.3. Prevention of Carbonylcyanide m-Chlorophenylhydrazone (CCCP)-Induced PLN Degradation by N-acetyl-l-cysteine (NAC) Pretreatment in PLN-Transfected HEK293 Cells
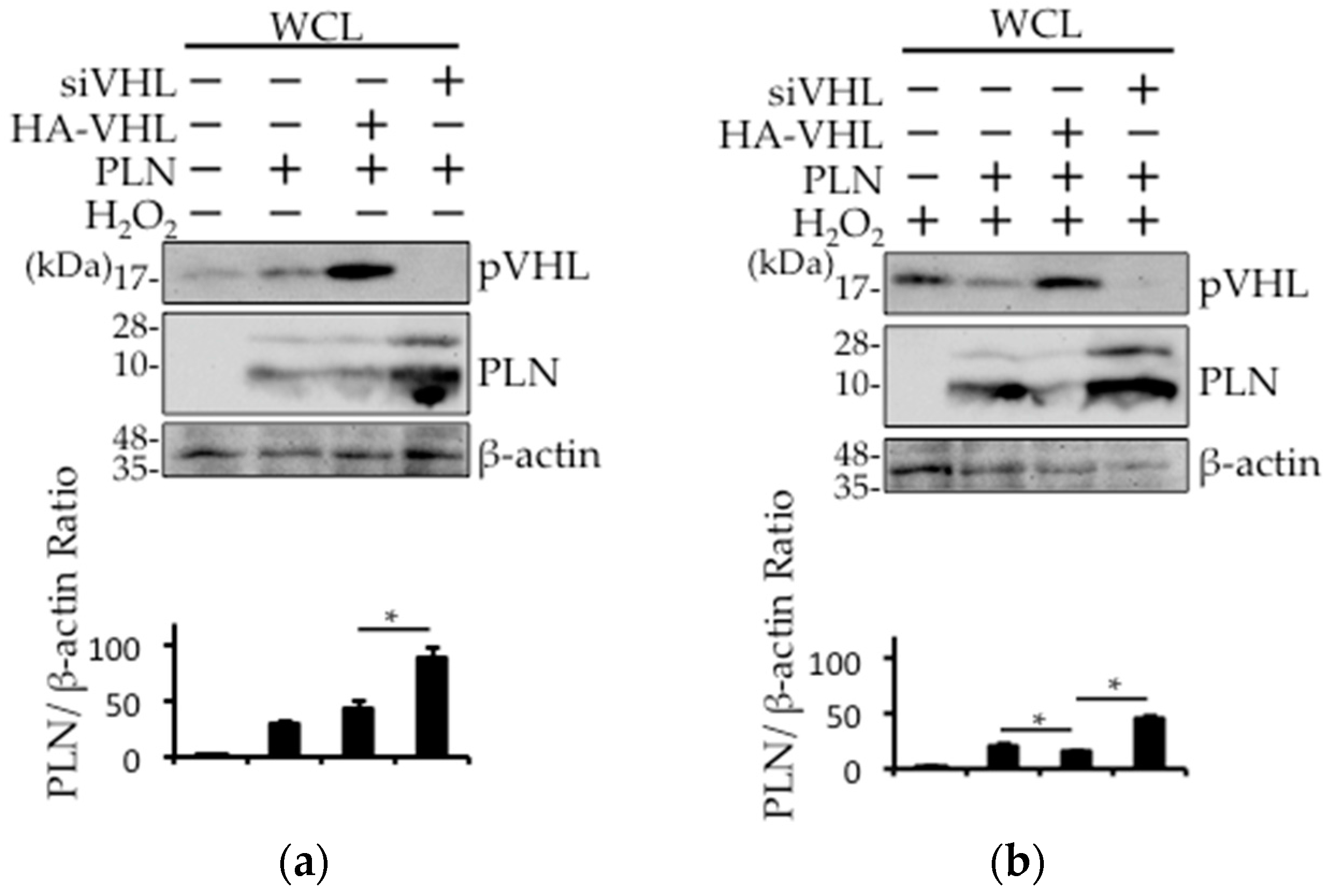
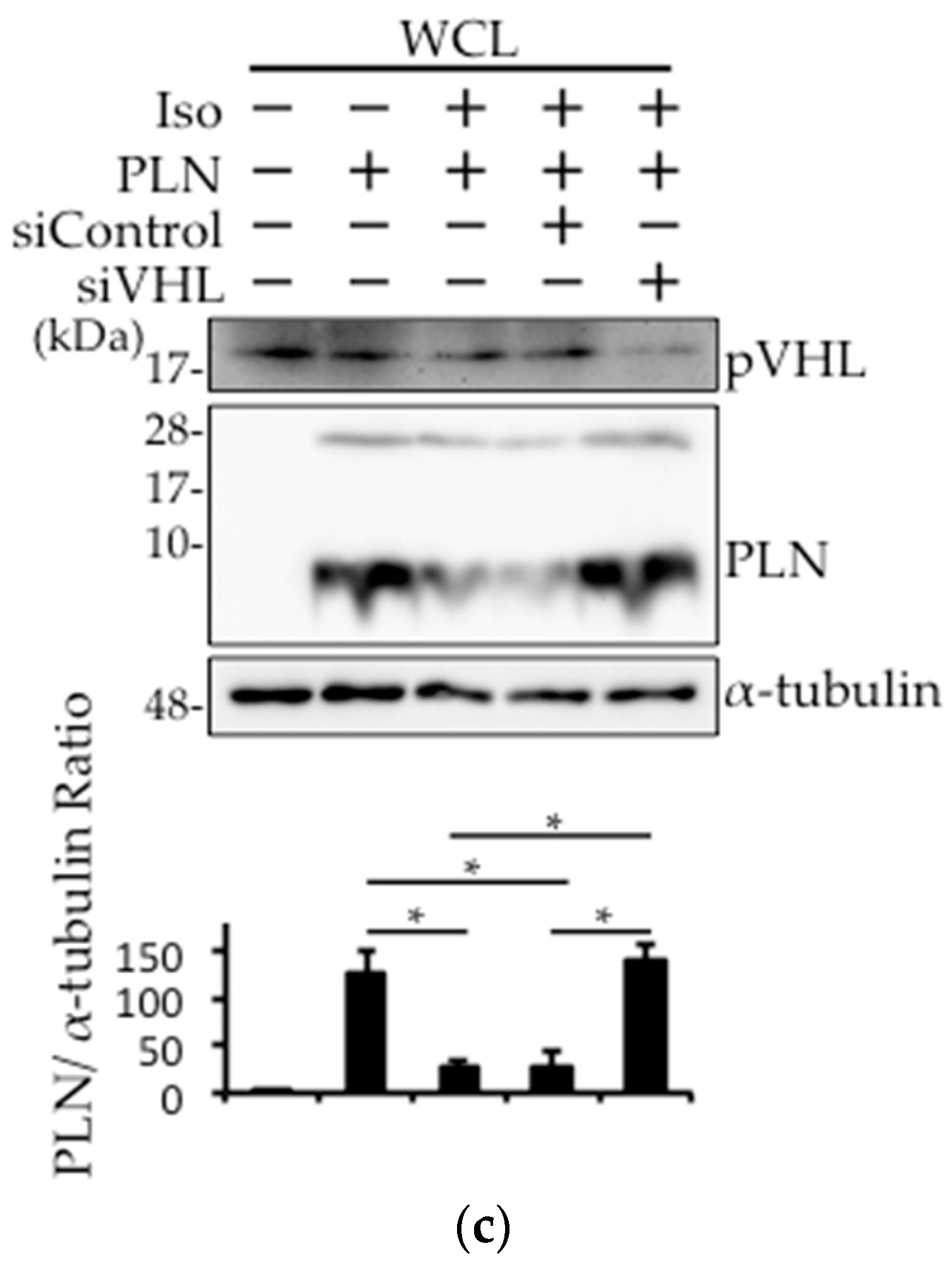
2.4. The Involvement of pVHL in H2O2- or Isoproterenol-Mediated PLN Degradation
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Animals
4.3. Cell Culture and Plasmid DNA or siRNA Transfection
4.4. Extraction of Mouse Heart Tissues
4.5. Western Blot Analysis
4.6. Co-Immunoprecipitation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SR | Sarcoplasmic reticulum |
| SERCA | Sarco(endo)plasmic reticulum Ca2+-ATPase |
| PLN | Phospholamban |
| WT | Wild-type |
| DCM | Dilated cardiomyopathy |
| CCCP | Carbonylcyanide m-chlorophenylhydrazone |
| NAC | N-acetyl-l-cysteine |
| UPS | Ubiquitin/proteasome system |
| VHL | Von Hippel–Lindau |
| HIF-1α | Hypoxia-inducible factor-1α |
| HACE1 | HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| BNP | Brain natriuretic peptide |
| WCL | Whole cell lysate |
| WB | Western blotting |
References
- Baehrecke, E.H. Autophagy: Dual roles in life and death? Nat. Rev. Mol. Cell Biol. 2005, 6, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. The ubiquitin-proteasome proteolytic pathway. Cell 1994, 79, 13–21. [Google Scholar] [CrossRef]
- Ciechanover, A. The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 1998, 17, 7151–7160. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.R. The ubiquitin-proteasome system in cardiac physiology and pathology. Am. J. Physiol. Heart Circ. Physiol. 2006, 291, H1–H19. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Robbins, J. Heart failure and protein quality control. Circ. Res. 2006, 99, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Day, S.M. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am. J. Physiol. Heart Circ. Physiol. 2013, 304, H1283–H1293. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W. Lipid droplets, lipophagy, and beyond. Biochim. Biophys. Acta 2016, 1861, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Li, H.H.; Willis, M.S.; Lockyer, P.; Miller, N.; McDonough, H.; Glass, D.J.; Patterson, C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J. Clin. Investig. 2007, 117, 3211–3223. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Guo, S.; Fan, Y.; Zhang, H.; Gu, D.; Li, H. Atrogin-1/MAFbx enhances simulated ischemia/reperfusion-induced apoptosis in cardiomyocytes through degradation of MAPK phosphatase-1 and sustained JNK activation. J. Biol. Chem. 2009, 284, 5488–5496. [Google Scholar] [CrossRef] [PubMed]
- Mearini, G.; Gedicke, C.; Schlossarek, S.; Witt, C.C.; Kramer, E.; Cao, P.; Gomes, M.D.; Lecker, S.H.; Labeit, S.; Willis, M.S.; et al. Atrogin-1 and MuRF1 regulate cardiac MyBP-C levels via different mechanisms. Cardiovasc. Res. 2010, 85, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chen, X.; Sharma, P.; Moon, M.; Sheftel, A.D.; Dawood, F.; Nghiem, M.P.; Wu, J.; Li, R.K.; Gramolini, A.O.; et al. HACE1-dependent protein degradation provides cardiac protection in response to haemodynamic stress. Nat. Commun. 2014, 5, 3430. [Google Scholar] [CrossRef] [PubMed]
- Zaglia, T.; Milan, G.; Ruhs, A.; Franzoso, M.; Bertaggia, E.; Pianca, N.; Carpi, A.; Carullo, P.; Pesce, P.; Sacerdoti, D.; et al. Atrogin-1 deficiency promotes cardiomyopathy and premature death via impaired autophagy. J. Clin. Investig. 2014, 124, 2410–2424. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.E.; Liao, J.Y.; He, J.; Schisler, J.C.; Newgard, C.B.; Drujan, D.; Glass, D.J.; Frederick, C.B.; Yoder, B.C.; Lalush, D.S.; et al. The ubiquitin ligase MuRF1 regulates PPARα activity in the heart by enhancing nuclear export via monoubiquitination. Mol. Cell. Endocrinol. 2015, 413, 36–48. [Google Scholar] [CrossRef] [PubMed]
- Teng, A.C.; Miyake, T.; Yokoe, S.; Zhang, L.; Rezende, L.M., Jr.; Sharma, P.; MacLennan, D.H.; Liu, P.P.; Gramolini, A.O. Metformin increases degradation of phospholamban via autophagy in cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, 7165–7170. [Google Scholar] [CrossRef] [PubMed]
- Lonser, R.R.; Glenn, G.M.; Walther, M.; Chew, E.Y.; Libutti, S.K.; Linehan, W.M.; Oldfield, E.H. von Hippel–Lindau disease. Lancet 2003, 361, 2059–2067. [Google Scholar] [CrossRef]
- Giordano, F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Investig. 2005, 115, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1, O2, and the 3 PHDs: How animal cells signal hypoxia to the nucleus. Cell 2001, 107, 1–3. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-α to the von Hippel–Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Ivan, M.; Kondo, K.; Yang, H.; Kim, W.; Valiando, J.; Ohh, M.; Salic, A.; Asara, J.M.; Lane, W.S.; Kaelin, W.G., Jr. HIFα targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science 2001, 292, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Yokoe, S.; Nakagawa, T.; Kojima, Y.; Higuchi, K.; Asahi, M. Indomethacin-induced intestinal epithelial cell damage is mediated by pVHL activation through the degradation of collagen I and HIF-1α. Biochem. Biophys. Res. Commun. 2015, 468, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Mason, S.; Liu, D.; Huang, Y.; Marks, C.; Hickey, R.; Jovin, I.S.; Pypaert, M.; Johnson, R.S.; Giordano, F.J. Hypoxia-inducible factor-dependent degeneration, failure, and malignant transformation of the heart in the absence of the von Hippel–Lindau protein. Mol. Cell. Biol. 2008, 28, 3790–3803. [Google Scholar] [CrossRef] [PubMed]
- Simmerman, H.K.; Jones, L.R. Phospholamban: Protein structure, mechanism of action, and role in cardiac function. Physiol. Rev. 1998, 78, 921–947. [Google Scholar] [PubMed]
- MacLennan, D.H.; Kranias, E.G. Phospholamban: A crucial regulator of cardiac contractility. Nat. Rev. Mol. Cell Biol. 2003, 4, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Yokoe, S.; Asahi, M. Phospholamban degradation is induced by phosphorylation-mediated ubiquitination and inhibited by interaction with cardiac type Sarco(endo)plasmic reticulum Ca2+-ATPase. Biochem. Biophys. Res. Commun. 2016, 472, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.P.; Kamisago, M.; Asahi, M.; Li, G.H.; Ahmad, F.; Mende, U.; Kranias, E.G.; MacLennan, D.H.; Seidman, J.G.; Seidman, C.E. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science 2003, 299, 1410–1413. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.Y.; Iwata, Y.; Arai, Y.; Komamura, K.; Wakabayashi, S. Activation of Na+/H+ exchanger 1 is sufficient to generate Ca2+ signals that induce cardiac hypertrophy and heart failure. Circ. Res. 2008, 103, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Weekes, J.; Morrison, K.; Mullen, A.; Wait, R.; Barton, P.; Dunn, M.J. Hyperubiquitination of proteins in dilated cardiomyopathy. Proteomics 2003, 3, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Birks, E.J.; Latif, N.; Enesa, K.; Folkvang, T.; Luong le, A.; Sarathchandra, P.; Khan, M.; Ovaa, H.; Terracciano, C.M.; Barton, P.J.; et al. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc. Res. 2008, 79, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Predmore, J.M.; Wang, P.; Davis, F.; Bartolone, S.; Westfall, M.V.; Dyke, D.B.; Pagani, F.; Powell, S.R.; Day, S.M. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation 2010, 121, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Baumgarten, A.; Bang, C.; Tschirner, A.; Engelmann, A.; Adams, V.; von Haehling, S.; Doehner, W.; Pregla, R.; Anker, M.S.; Blecharz, K.; et al. TWIST1 regulates the activity of ubiquitin proteasome system via the miR-199/214 cluster in human end-stage dilated cardiomyopathy. Int. J. Cardiol. 2013, 168, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Ide, T.; Tsutsui, H.; Kinugawa, S.; Utsumi, H.; Kang, D.; Hattori, N.; Uchida, K.; Arimura, K.; Egashira, K.; Takeshita, A. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ. Res. 1999, 85, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Ide, T.; Tsutsui, H.; Kinugawa, S.; Suematsu, N.; Hayashidani, S.; Ichikawa, K.; Utsumi, H.; Machida, Y.; Egashira, K.; Takeshita, A. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ. Res. 2000, 86, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Hon, W.C.; Wilson, M.I.; Harlos, K.; Claridge, T.D.; Schofield, C.J.; Pugh, C.W.; Maxwell, P.H.; Ratcliffe, P.J.; Stuart, D.I.; Jones, E.Y. Structural basis for the recognition of hydroxyproline in HIF-1 α by pVHL. Nature 2002, 417, 975–978. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yokoe, S.; Asahi, M. Phospholamban Is Downregulated by pVHL-Mediated Degradation through Oxidative Stress in Failing Heart. Int. J. Mol. Sci. 2017, 18, 2232. https://doi.org/10.3390/ijms18112232
Yokoe S, Asahi M. Phospholamban Is Downregulated by pVHL-Mediated Degradation through Oxidative Stress in Failing Heart. International Journal of Molecular Sciences. 2017; 18(11):2232. https://doi.org/10.3390/ijms18112232
Chicago/Turabian StyleYokoe, Shunichi, and Michio Asahi. 2017. "Phospholamban Is Downregulated by pVHL-Mediated Degradation through Oxidative Stress in Failing Heart" International Journal of Molecular Sciences 18, no. 11: 2232. https://doi.org/10.3390/ijms18112232



