Identification of Two Cytochrome Monooxygenase P450 Genes, CYP321A7 and CYP321A9, from the Tobacco Cutworm Moth (Spodoptera Litura) and Their Expression in Response to Plant Allelochemicals
Abstract
:1. Introduction
2. Results
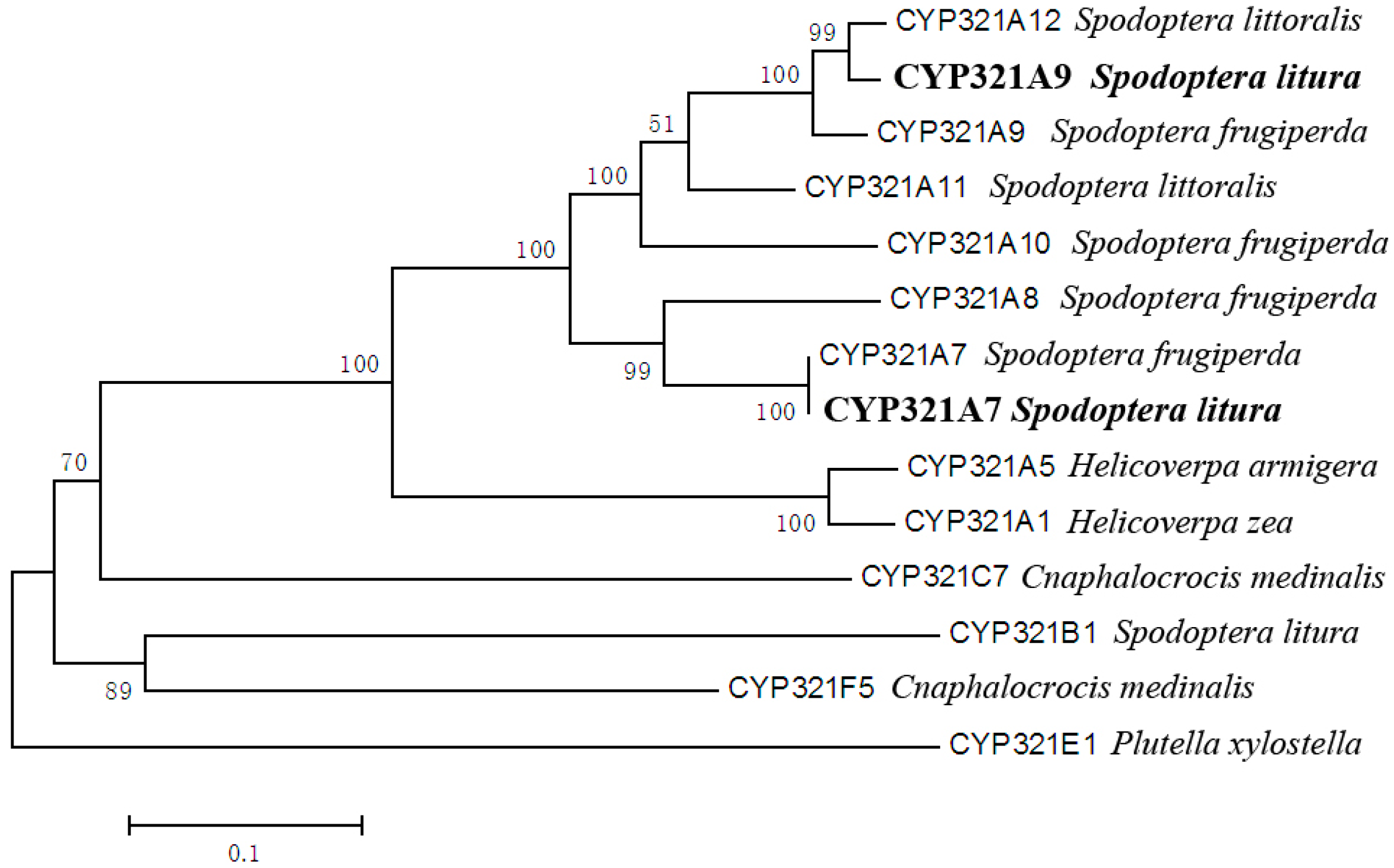
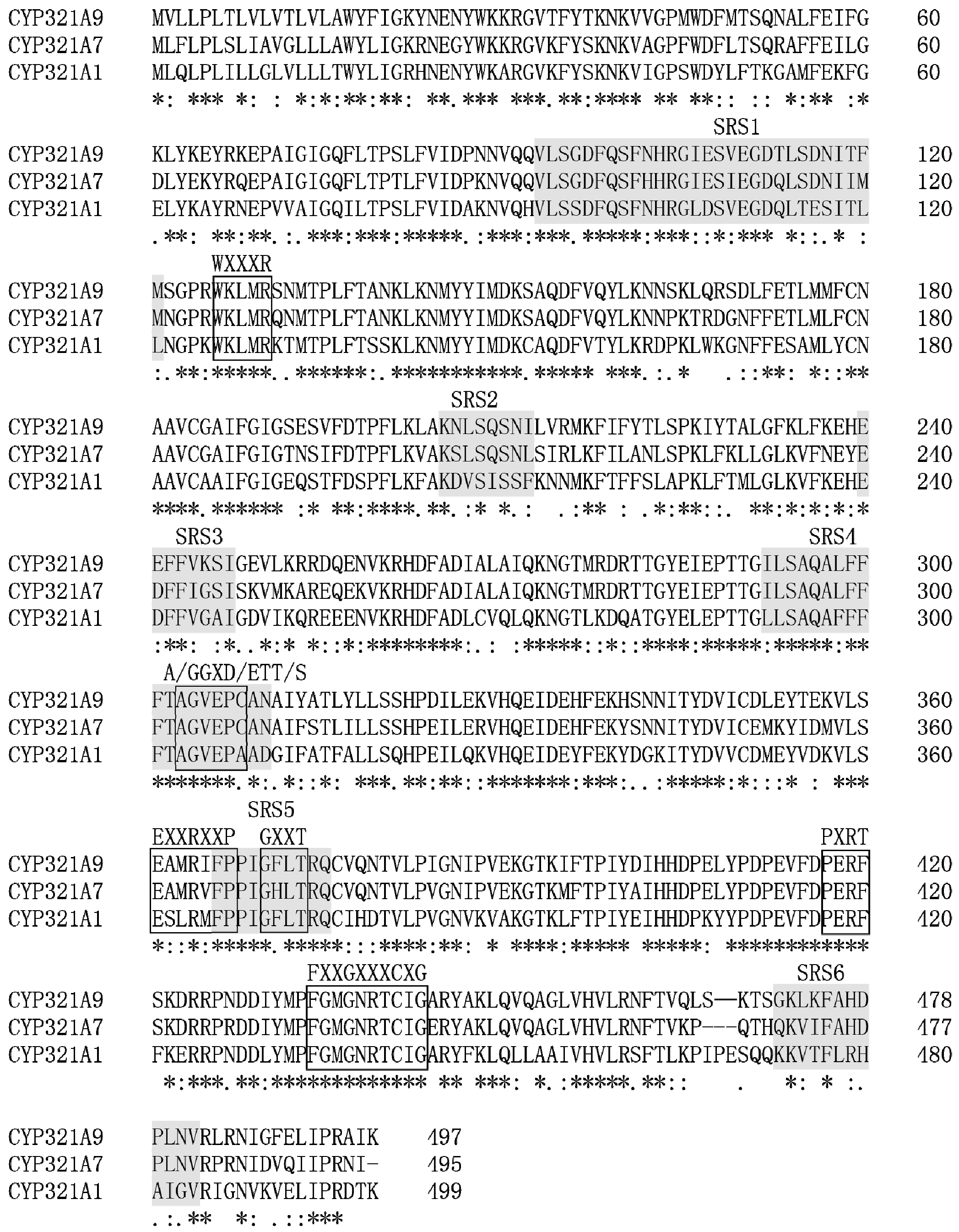
2.1. Identification of Two Novel Cytochrome P450 Genes Expressed in S. litura
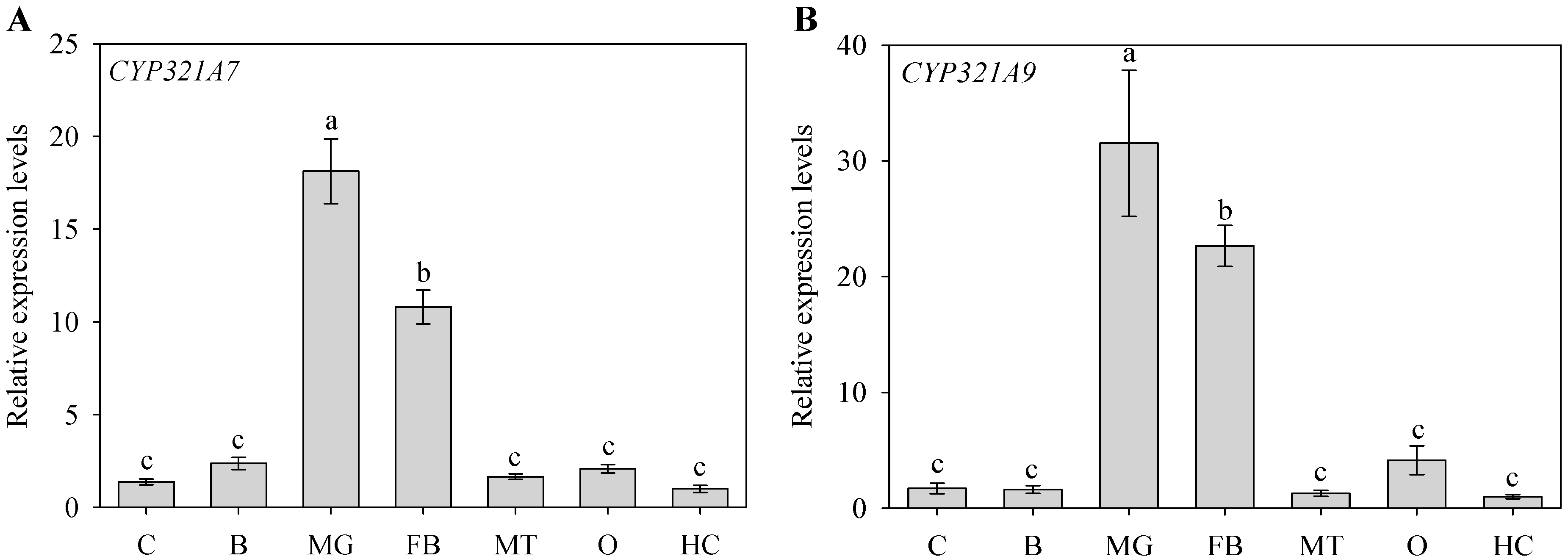
2.2. Tissue-Dependent Expression Pattern of CYP321A7 and CYP321A9
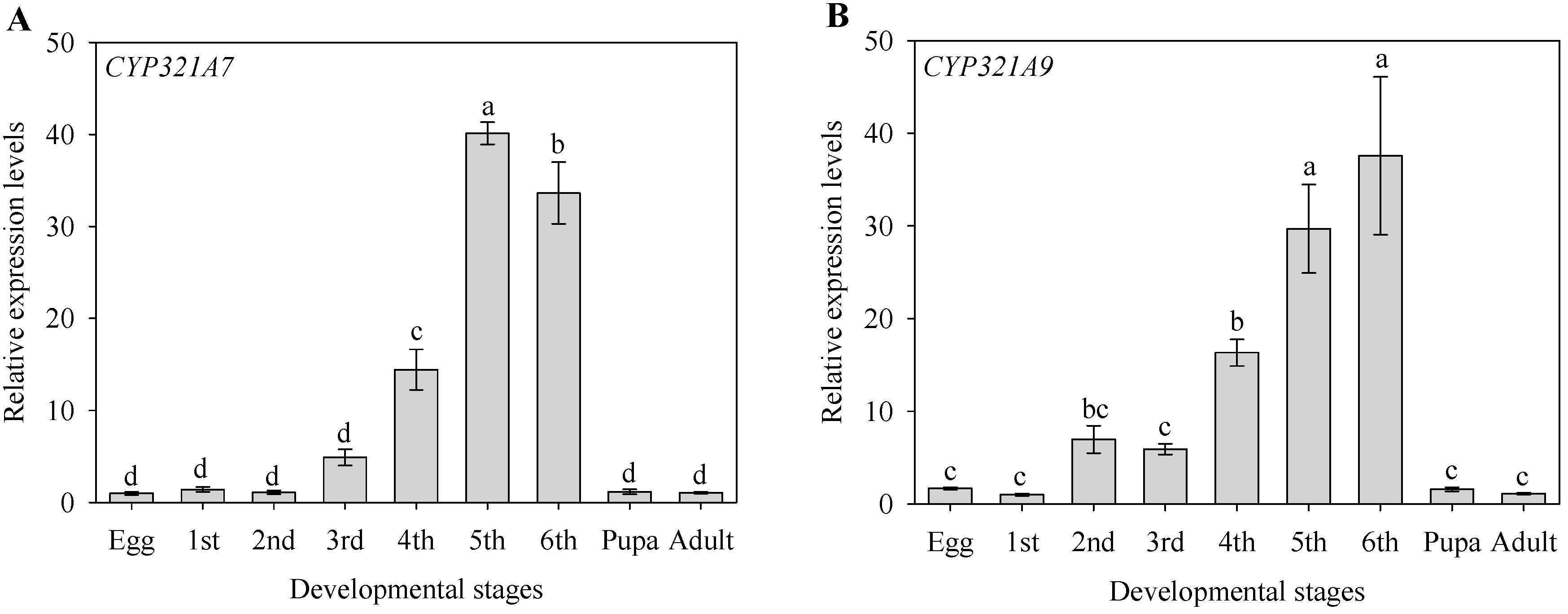
2.3. Stage-Dependent Expression Pattern of CYP321A7 and CYP321A9
2.4. Altered Expression of CYP321A7 and CYP321A9 in Response to Plant Allelochemcials
3. Discussion
4. Experimental Section
4.1. Insect Rearing
4.2. Identification of CYP321A7 and CYP321A9 by Whole Trancriptome Analysis
4.3. Cloning of CYP321A7 and CYP321A9
4.4. Bioinformatic Analysis
4.5. Plant Allelochemicals
4.6. Expression of CYP321A7 and CYP321A9 in Different Tissues
4.7. Stage-Dependent Expression Pattern of CYP321A7 and CYP321A9
4.8. Effects of Different Plant Allelochemicals on CYP321A7 and CYP321A9 Expression
4.9. RT-qPCR Analysis
4.10. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xue, H.; Pang, Y.H.; Li, Q.L.; Liu, T.X. Effects of four host plants on susceptibility of Spodoptera litura larvae to five insecticides and activities of detoxification. Pest Manag. Sci. 2010, 66, 1273–1279. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Xia, Q.Q.; Baerson, S.R.; Ren, Y.; Wang, J.; Su, Y.J.; Zheng, S.C.; Zeng, R.S. A novel cytochrome P450 CYP6AB14 gene in Spodoptera litura (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J. Insect Physiol. 2015, 75, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Feyereisen, R. Evolution of insect P450. Biochem. Soc. Trans. 2006, 34, 1252–1255. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 2007, 52, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Giraudo, M.; Hilliou, F.; Fricaux, T.; Audant, P.; Feyereisen, R.; Le, G.G. Cytochrome P450s from the fall armyworm (Spodoptera frugiperda): Responses to plant allelochemicals and pesticides. Insect Mol. Biol. 2015, 24, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Li, J.; Staehelin, C.; Xin, X.W.; Su, Y.J.; Zeng, R.S. Expression analysis of two P450 monooxygenase genes of the tobacco cutworm moth (Spodoptera litura) at different developmental stages and in response to plant allelochemicals. J. Chem. Ecol. 2015, 41, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Ma, K.S.; Liang, P.Z.; Chen, X.W.; Liu, Y.; Gao, X.W. Transcriptional responses of detoxification genes to four plant allelochemicals in Aphis gossypii. J. Econ. Entomol. 2017, 110, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Pottier, M.A.; Bozzolan, F.; Chertemps, T.; Jacquin-Joly, E.; Lalouette, L.; Siaussat, D.; Maïbèche-Coisne, M. Cytochrome P450s and cytochrome P450 reductase in the olfactory organ of the cotton leafworm Spodoptera littoralis. Insect Mol. Biol. 2012, 21, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Rupasinghe, S.G.; Wen, Z.; Chiu, T.L.; Schuler, M.A. Helicoverpa zea CYP6B8 and CYP321A1: Different molecular solutions to the problem of metabolizing plant toxins and insecticides. Protein Eng. Des. Sel. 2007, 20, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.S.; Wen, Z.; Niu, G.; Schuler, M.A.; Berenbaum, M.R. Allelochemical induction of cytochrome P450 monooxygenases and amelioration of xenobiotic toxicity in Helicoverpa zea. J. Chem. Ecol. 2007, 33, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Berenbaum, M.R.; Schuler, M.A. Plant allelochemicals differentially regulate Helicoverpa zea cytochrome P450 genes. Insect Mol. Biol. 2002, 11, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.M.; Zeng, R.S.; Niu, G.; Berenbaum, M.R.; Schuler, M.A. Ecological significance of induction of broad-substrate cytochrome P450s by natural and synthetic Inducers in Helicoverpa zea. J. Chem. Ecol. 2009, 35, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liang, P.; Gao, X.; Shi, X. Induction of the cytochrome P450 activity by plant allelochemicals in the cotton bollworm, Helicoverpa armigera (Hübner). Pestic. Biochem. Physiol. 2006, 84, 127–134. [Google Scholar] [CrossRef]
- Petersen, R.A.; Zangerl, A.R.; Berenbaum, M.R.; Schuler, M.A. Expression of CYP6B1 and CYP6B3 cytochrome P450 monooxygenases and furanocoumarin metabolism in different tissues of Papilio polyxenes (Lepidoptera: Papilionidae). Insect Biochem. Mol. Biol. 2001, 31, 679–690. [Google Scholar] [CrossRef]
- Zhang, Y.E.; Ma, H.J.; Feng, D.D.; Lai, X.F.; Chen, Z.M.; Xu, M.Y.; Yu, Q.Y.; Zhang, Z. Induction of detoxification enzymes by quercetin in the Silkworm. J. Econ. Entomol. 2012, 105, 1034–1042. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.R.; Scott, I.M.; Sims, S.; Trudeau, V.L.; Arnason, J.T. Gene expression profiles of Drosophila melanogaster exposed to an insecticidal extract of Piper nigrum. J. Agric. Food Chem. 2006, 54, 1289–1295. [Google Scholar] [CrossRef] [PubMed]
- David, J.P.; Boyer, S.; Mesneau, A.; Ball, A.; Ranson, H.; Dauphinvillemant, C. Involvement of cytochrome P450 monooxygenases in the response of mosquito larvae to dietary plant xenobiotics. Insect Biochem. Mol. Bol. 2006, 36, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, S.; Dean, E.D.; Lam, V.; Ganguly, R. Induction of two cytochrome P450 genes, Cyp6a2 and Cyp6a8, of Drosophila melanogaster by caffeine in adult flies and in cell culture. Gene 2006, 377, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Staehelin, C.; Xia, Q.Q.; Su, Y.J.; Zeng, R.S. Identification and characterization of CYP9A40 from the tobacco cutworm moth (Spodoptera litura), a cytochrome P450 gene induced by plant allelochemicals and insecticides. Int. J. Mol. Sci. 2015, 16, 22606–22620. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.L.; Zhu-Salzman, K.; Baerson, S.R.; Xin, X.W.; Li, J.; Su, Y.J.; Zeng, R.S. Identification of a novel cytochrome p450 CYP321B1 gene from tobacco cutworm (Spodoptera litura) and RNA interference to evaluate its role in commonly used insecticides. Insect Sci. 2017, 24, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Després, L.; David, J.P.; Gallet, C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol. Evol. 2007, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Sasabe, M.; Wen, Z.; Berenbaum, M.R.; Schuler, M.A. Molecular analysis of CYP321A1, a novel cytochrome P450 involved in metabolism of plant allelochemicals (furanocoumarins) and insecticides (cypermethrin) in Helicoverpa zea. Gene 2004, 338, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Schuler, M.A.; Berenbaum, M.R. Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature 2002, 419, 712–715. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.S.; Li, F.; Liu, Y.; Liang, P.Z.; Chen, X.W.; Gao, X.W. Identification of microRNAs and their response to the stress of plant allelochemicals in Aphis gossypii (Hemiptera: Aphididae). BMC Mol. Biol. 2017, 18, 5. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, O. Substrate recognition sites in cytochrome P450 family 2 (CYP2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J. Biol. Chem. 1992, 267, 83–90. [Google Scholar] [PubMed]
- Zhu, F.; Parthasarathy, R.; Bai, H.; Woithe, K.; Kaussmann, M.; Nauen, R.; Harrison, D.A.; Palli, S.R. A brain-specific cytochrome P450 responsible for the majority of deltamethrin resistance in the QTC279 strain of Tribolium castaneum. Proc. Natl. Acad. Sci. USA 2010, 107, 8557–8562. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.S.; Wen, Z.; Niu, G.; Schuler, M.A.; Berenbaum, M.R. Enhanced toxicity and induction of cytochrome P450s suggest a cost of “eavesdropping” in a multitrophic interaction. J. Chem. Ecol. 2009, 35, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Wybouw, N.; Rombauts, S.; Menten, B.; Vontas, J.; Grbic, M.; Clark, R.M.; Feyereisen, R.; Van, L.T. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. USA 2013, 110, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, M.; Palli, S.R. Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 2017, 83, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.A. P450s in plant-insect interactions. BBA-Proteins Proteom. 2011, 1814, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.M.; Berenbaum, M.R.; Schuler, M.A. Inhibition of CYP6B1-mediated detoxification of xanthotoxin by plant allelochemicals in the black swallowtail (Papilio polyxenes). J. Chem. Ecol. 2006, 32, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ma, C.; Li, M.; Sheng, C.; Liu, H.; Qiu, X. CYP9A12 and CYP9A17 in the cotton bollworm, Helicoverpa armigera: Sequence similarity, expression profile and xenobiotic response. Pest Manag. Sci. 2010, 66, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.J.; Li, G.H.; Pang, Y. A simple artificial diet for mass rearing of some noctuid species. Entomol. Knowl. 2000, 37, 8–10. [Google Scholar]
- Ahmad, M.; Ghaffar, A.; Rafiq, M. Host plants of leaf worm, Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae) in Pakistan. Asian J. Agric. Biol. 2013, 1, 23–28. [Google Scholar]
- Zeng, R.S.; Wen, Z.; Niu, G.; Berenbaum, M.R. Aflatoxin B1: Toxicity, bioactivation and detoxification in the polyphagous caterpillar Trichoplusia ni. Insect Sci. 2013, 20, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.-L.; He, Y.-N.; Staehelin, C.; Liu, S.-W.; Su, Y.-J.; Zhang, J.-E. Identification of Two Cytochrome Monooxygenase P450 Genes, CYP321A7 and CYP321A9, from the Tobacco Cutworm Moth (Spodoptera Litura) and Their Expression in Response to Plant Allelochemicals. Int. J. Mol. Sci. 2017, 18, 2278. https://doi.org/10.3390/ijms18112278
Wang R-L, He Y-N, Staehelin C, Liu S-W, Su Y-J, Zhang J-E. Identification of Two Cytochrome Monooxygenase P450 Genes, CYP321A7 and CYP321A9, from the Tobacco Cutworm Moth (Spodoptera Litura) and Their Expression in Response to Plant Allelochemicals. International Journal of Molecular Sciences. 2017; 18(11):2278. https://doi.org/10.3390/ijms18112278
Chicago/Turabian StyleWang, Rui-Long, Ya-Nan He, Christian Staehelin, Shi-Wei Liu, Yi-Juan Su, and Jia-En Zhang. 2017. "Identification of Two Cytochrome Monooxygenase P450 Genes, CYP321A7 and CYP321A9, from the Tobacco Cutworm Moth (Spodoptera Litura) and Their Expression in Response to Plant Allelochemicals" International Journal of Molecular Sciences 18, no. 11: 2278. https://doi.org/10.3390/ijms18112278




