Efficient Generation of Genome-Modified Mice Using Campylobacter jejuni-Derived CRISPR/Cas
Abstract
:1. Introduction
2. Results
2.1. Examination of PAM Sequences Applicable to Mouse Zygotes
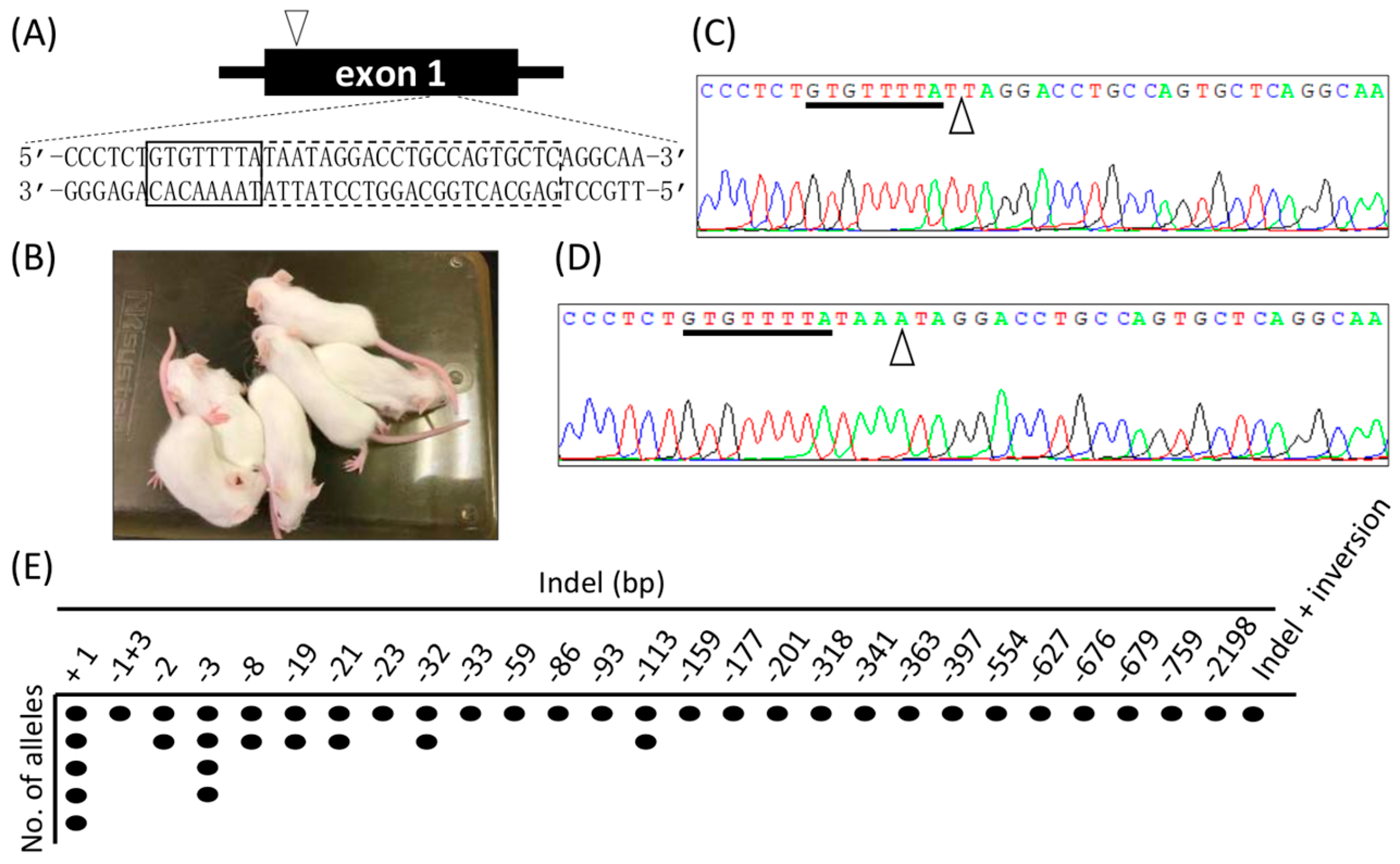
2.2. Generation of KO Mice Using Cj-CRISPR/Cas
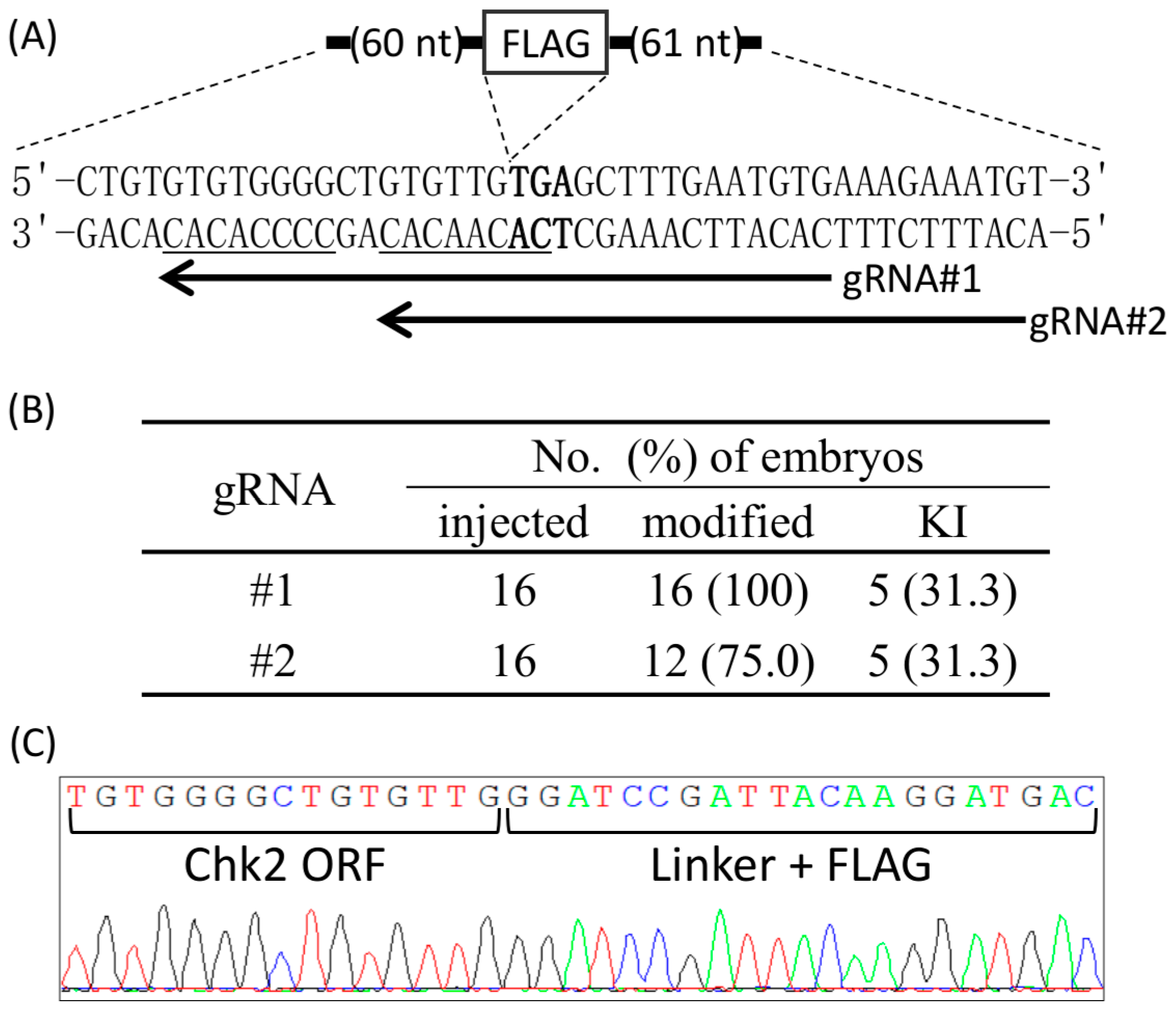
2.3. Zygote-Mediated KI by Cj-CRISPR/Cas
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Construction of Cas9-and gRNA-plasmid DNA
4.3. DNA Transfection and Immunoblotting to Validate Cj-Cas9 Expression
4.4. In Vitro Transcription of Cas9 mRNA and gRNAs
4.5. In Vitro Fertilization
4.6. Microinjection and Embryo-Transfer
4.7. Detection of Induced Mutations
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeat |
| Cas | CRISPR-Associated |
| PAM | protospacer adjacent motif |
| gRNA | guide-RNA |
| DSB | DNA double-strand break |
| NHEJ | non-homologous end-joining |
| Sp | Streptococcus pyogenes |
| Cj | Campylobacter jejuni |
| KO | knockout |
| KI | knock-in |
| ssODN | single-strand oligodeoxynucleotide |
| Chk2 | checkpoint kinase 2 |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | fetal bovine serum |
| eCG | equine chorionic gonadotropin |
| hCG | human chorionic gonadotropin |
References
- Meyer, M.; de Angelis, M.H.; Wurst, W.; Kühn, R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 15022–15026. [Google Scholar] [CrossRef] [PubMed]
- Geurts, A.M.; Cost, G.J.; Freyvert, Y.; Zeitler, B.; Miller, J.C.; Choi, V.M.; Jenkins, S.S.; Wood, A.; Cui, X.; Meng, X.; et al. Knockout rats via embryo microinjection of zinc-finger nucleases. Science 2009, 325, 433. [Google Scholar] [CrossRef] [PubMed]
- Sung, Y.H.; Baek, I.J.; Kim, D.H.; Jeon, J.; Lee, J.; Lee, K.; Jeong, D.; Kim, J.S.; Lee, H.W. Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 2013, 31, 23–24. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Zhang, J.; Wu, H.; Wang, J.; Ma, K.; Li, Z.; Zhang, X.; Zhang, P.; Huang, X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013, 23, 720–723. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xu, J.; Zhu, T.; Fan, J.; Lai, L.; Zhang, J.; Chen, Y.E. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J. Mol. Cell Biol. 2014, 6, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Hai, T.; Teng, F.; Guo, R.; Li, W.; Zhou, Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014, 24, 372–375. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Fujii, W.; Kawasaki, K.; Sugiura, K.; Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013, 41, e187. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Fonfara, I.; Le Rhun, A.; Chylinski, K.; Makarova, K.S.; Lécrivain, A.L.; Bzdrenga, J.; Koonin, E.V.; Charpentier, E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 2577–2590. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.J.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and Engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Fujii, W.; Kakuta, S.; Yoshioka, S.; Kyuwa, S.; Sugiura, K.; Naito, K. Zygote-mediated generation of genome-modified mice using Streptococcus thermophilus 1-derived CRISPR/Cas system. Biochem. Biophys. Res. Commun. 2016, 477, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Watanabe, Y.; Gootenberg, J.S.; Hirano, H.; Ran, F.A.; Nakane, T.; Ishitani, R.; Zhang, F.; Nishimasu, H.; Nureki, O. Crystal Structure of the Minimal Cas9 from Campylobacter jejuni Reveals the Molecular Diversity in the CRISPR-Cas9 Systems. Mol. Cell 2017, 65, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Fujii, W. Generation of Knock-in Mouse by Genome Editing. Methods Mol. Biol. 2017, 1630, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Fujii, W.; Kano, K.; Sugiura, K.; Naito, K. Repeatable construction method for engineered zinc finger nuclease based on overlap extension PCR and TA-cloning. PLoS ONE 2013, 8, e59801. [Google Scholar] [CrossRef] [PubMed]
- Iseli, C.; Ambrosini, G.; Bucher, P.; Jongeneel, C.V. Indexing strategies for rapid searches of short words in genome sequences. PLoS ONE 2007, 2, e579. [Google Scholar] [CrossRef] [PubMed]
| gRNA * | PAM Sequence | Mutated/Total | |||||
|---|---|---|---|---|---|---|---|
| 1st-3rd | 4th | 5th | 6th | 7th | 8th | (%) | |
| Reported PAM | |||||||
| 5′-NNNNACA [14] | NNN | all | A | C | A | all | − |
| 5′-NNNNRYAC [18] | NNN | all | A/G | C/T | A | C | − |
| 5′-NNNVRYM [19] | NNN | no T | A/G | C/T | A/C | all | − |
| common in all | NNN | no T | A | C | A | C | |
| T1 | TAA | A | A | C | A | C | 16/16 (100) |
| T2 | TGT | G | A | C | A | C | 16/16 (100) |
| R1 | TGC | A | A | C | A | C | 16/16 (100) |
| 5th and/or 6th change | NNN | no T | G | T | A | C | |
| T3 | GGC | A | A | T | A | C | 12/16 (75.0) |
| T4 | TGA | A | G | T | A | C | 7/16 (43.8) |
| R2 | GGC | C | G | C | A | C | 13/16 (81.3) |
| 7th change | NNN | no T | A | C | C | C | |
| T5 | ATC | C | A | C | C | C | 0/16 (0) |
| T6 | CAT | C | A | C | C | C | 0/16 (0) |
| T7 (also 5th) | CCT | G | G | C | C | C | 0/16 (0) |
| 8th change | NNN | no T | A | C | A | no C | |
| T8 | AAA | C | A | C | A | G | 0/16 (0) |
| T9 | AAG | A | A | C | A | T | 0/16 (0) |
| 4th change | NNN | T | A | C | A | C | |
| T10 | TTT | T | A | C | A | C | 0/16 (0) |
| T11 (also 5th) | CAG | T | G | C | A | C | 0/16 (0) |
| T12 (also 5th) | TGG | T | G | C | A | C | 0/16 (0) |
| T13 (also 5/6th) | AAT | T | G | T | A | C | 0/16 (0) |
| R3 | TTT | T | A | C | A | C | 0/16 (0) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fujii, W.; Ikeda, A.; Sugiura, K.; Naito, K. Efficient Generation of Genome-Modified Mice Using Campylobacter jejuni-Derived CRISPR/Cas. Int. J. Mol. Sci. 2017, 18, 2286. https://doi.org/10.3390/ijms18112286
Fujii W, Ikeda A, Sugiura K, Naito K. Efficient Generation of Genome-Modified Mice Using Campylobacter jejuni-Derived CRISPR/Cas. International Journal of Molecular Sciences. 2017; 18(11):2286. https://doi.org/10.3390/ijms18112286
Chicago/Turabian StyleFujii, Wataru, Arisa Ikeda, Koji Sugiura, and Kunihiko Naito. 2017. "Efficient Generation of Genome-Modified Mice Using Campylobacter jejuni-Derived CRISPR/Cas" International Journal of Molecular Sciences 18, no. 11: 2286. https://doi.org/10.3390/ijms18112286