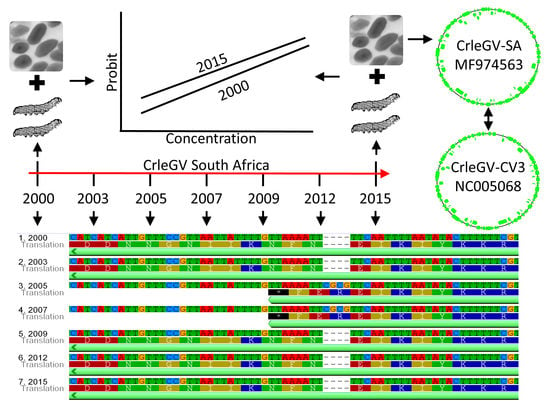
Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide
Abstract
:1. Introduction
2. Results
2.1. Comparison of CrleGV-SA Genome Sequences Across a 15-Year Period
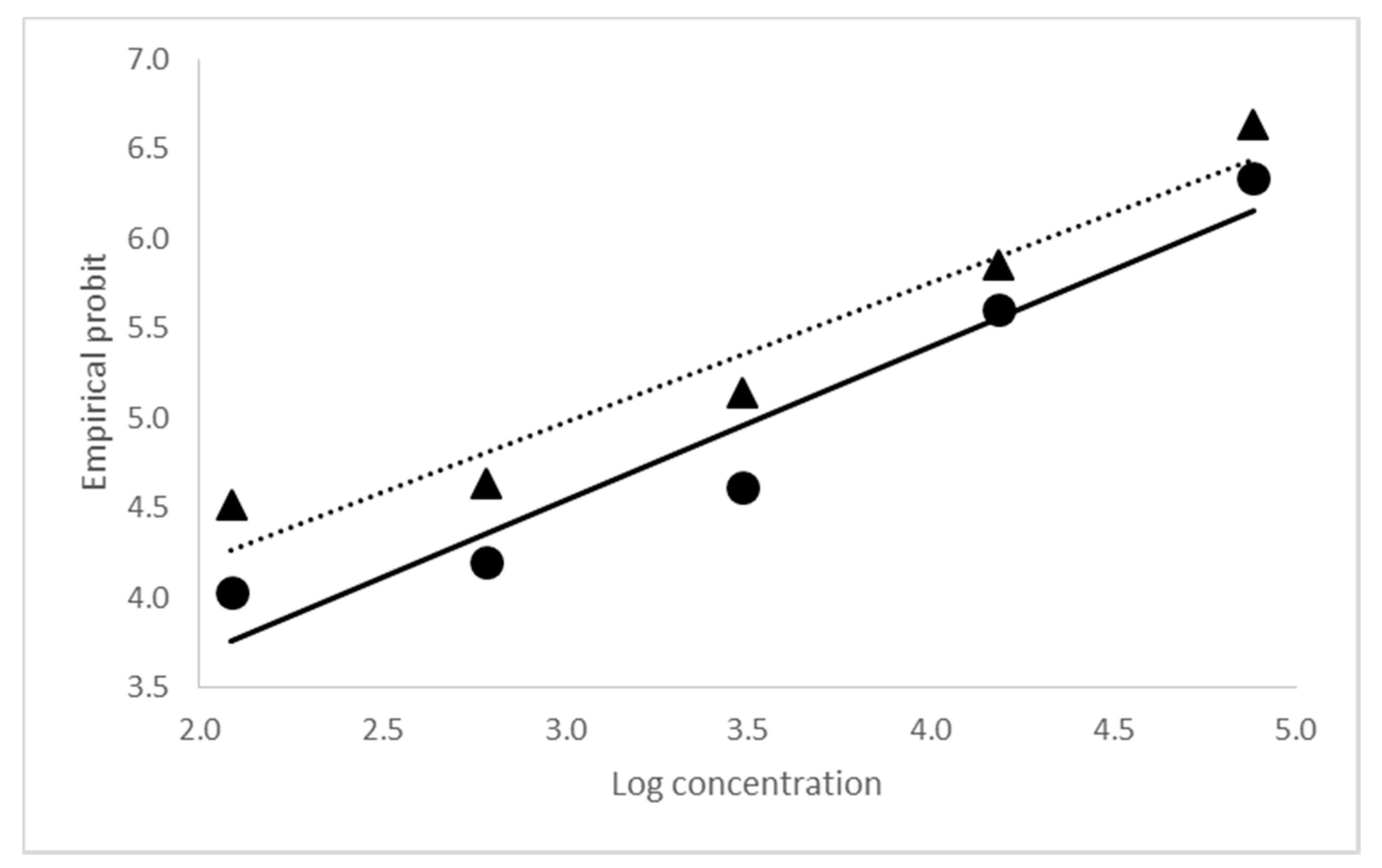
2.2. Biological Activity of CrleGV-SA 2000 and 2015
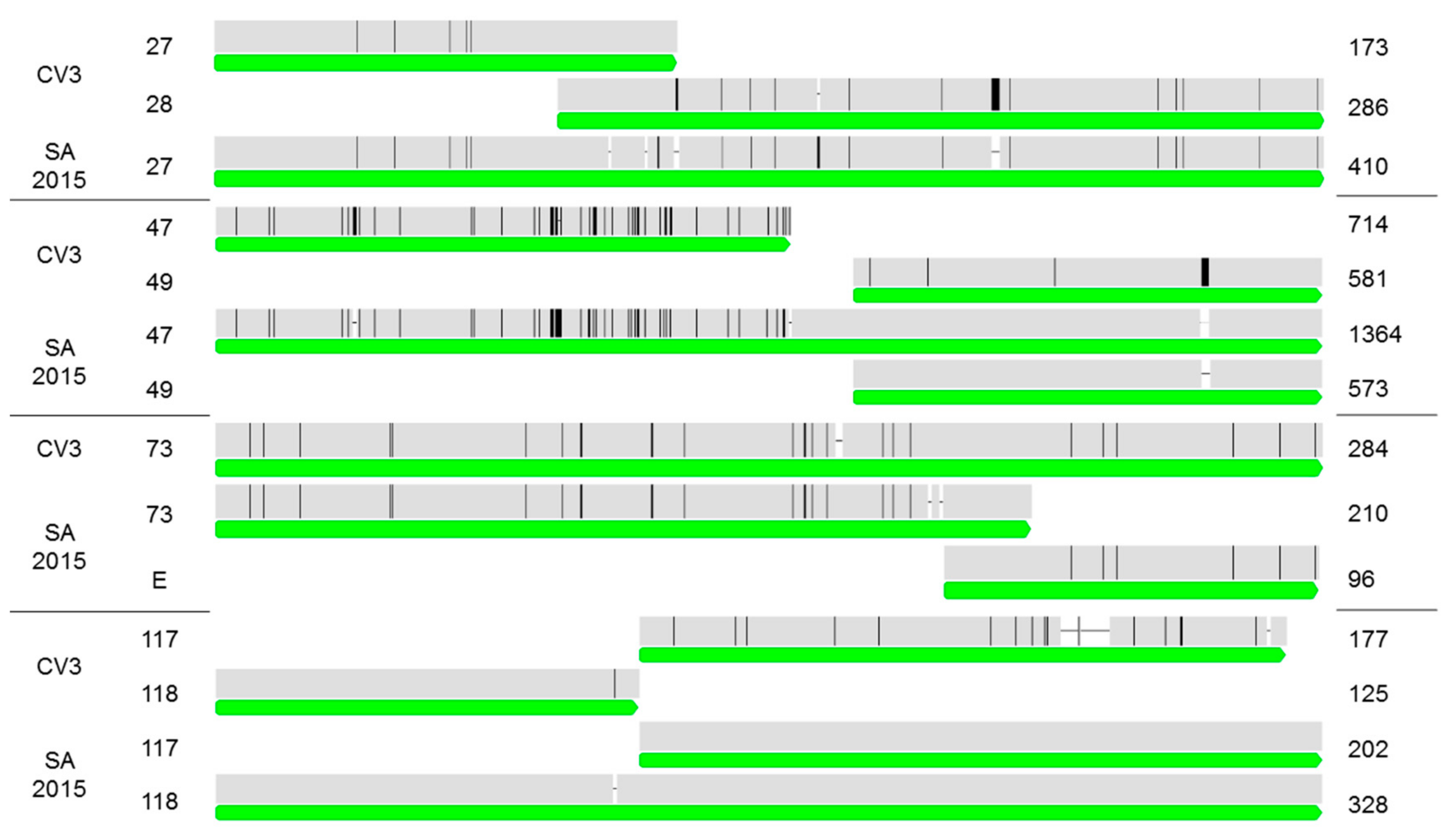
2.3. Comparison of the CrleGV-SA 2015 Genome Sequence to CrleGV-CV3
3. Discussion
4. Materials and Methods
4.1. Host Material
4.2. Virus Inoculum
4.3. Genomic DNA Extraction
4.4. CrleGV-SA Genome Sequencing and Analysis
4.5. Biological Assays
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Newton, P.J. False codling moth Cryptophlebia leucotreta (Meyrick). In Citrus Pests in the Republic of South Africa; ARC: Nelspruit, South Africa, 1998; pp. 194–200. [Google Scholar]
- Moore, S.D.; Kirkman, W.; Stephen, P.R.; Albertyn, S.; Love, C.N.; Grout, T.G.; Hattingh, V. Development of an improved postharvest cold treatment for Thaumatotibia leucotreta (Meyrick) (Lepidoptera: Tortricidae). Postharvest Biol. Technol. 2017, 125, 188–195. [Google Scholar] [CrossRef]
- Knox, C.; Moore, S.D.; Luke, G.A.; Hill, M.P. Baculovirus-based strategies for the management of insect pests: A focus on development and application in South Africa. Biocontrol Sci. Technol. 2015, 25, 1–20. [Google Scholar] [CrossRef]
- Moore, S.D.; Hattingh, V. A review of current pre-harvest control options for false codling moth in citrus in southern Africa. S. Afr. Fruit J. 2012, 11, 82–85. [Google Scholar]
- Angélini, A.; Amargier, A.; Vandamme, P.; Duthoit, J.L. Une virose á granules chez le lepidoptére Argyroploce leucotreta. Coton Fibres Trop. 1965, 20, 277–282. [Google Scholar]
- Mück, O. Biologie, Verhalten und Wirtschaftliche Bedeutung von Parasiten Schädlicher Lepidopteren auf den Kapverden; Bauer: Exeter, NH, USA, 1985. [Google Scholar]
- Jehle, J.A.; Backhaus, H.; Fritsch, E.; Huber, J. Physical map of the Cryptophlebia leucotreta granulosis virus genome and its relationship to the genome of Cydia pomonella granulosis virus. J. Gen. Virol. 1992, 73, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, E. Das Granulosevirus des Falschen Apfelwicklers, Cryptophlebia leucotreta (Meyrick). Ph.D. Thesis, Technische Universität, Darmstadt, Germany, December 1989. [Google Scholar]
- Lange, M.; Jehle, J.A. The genome of the Cryptophlebia leucotreta granulovirus. Virology 2003, 317, 220–236. [Google Scholar] [CrossRef]
- Singh, S.; Moore, S.; Spillings, B.; Hendry, D. South African isolate of Cryptophlebia leucotreta granulovirus. J. Invertebr. Pathol. 2003, 83, 249–252. [Google Scholar] [CrossRef]
- Moore, S.D.; Kirkman, W.; Richards, G.I.; Stephen, P.R. The Cryptophlebia leucotreta granulovirus—10 years of commercial field use. Viruses 2015, 7, 1284–1312. [Google Scholar] [CrossRef] [PubMed]
- Grzywacz, D.; Moore, S. Production, Formulation, and Bioassay of Baculoviruses for Pest Control. In Microbial Control of Insect and Mite Pests; Academic Press: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Herniou, E.A.; Jehle, J.A. Baculovirus phylogeny and evolution. Curr. Drug Targets 2007, 8, 1043–1050. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Debrah, J.K.; Hill, M.P.; Knox, C.; Moore, S.D. Overcrowding of false codling moth, Thaumatotibia leucotreta (Meyrick) leads to the isolation of five new Cryptophlebia leucotreta granulovirus (CrleGV-SA) isolates. J. Invertebr. Pathol. 2013, 112, 219–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. A novel binary mixture of Helicoverpa armigera single nucleopolyhedrovirus genotypic variants has improved insecticidal characteristics for control of cotton bollworms. Appl. Environ. Microbiol. 2015, 81, 3984–3993. [Google Scholar] [CrossRef] [PubMed]
- Opoku-Debrah, J.K.; Hill, M.P.; Knox, C.; Moore, S.D. Heterogeneity in virulence relationships between Cryptophlebia leucotreta. BioControl 2016, 61, 449–459. [Google Scholar] [CrossRef]
- Erlandson, M.A. Genetic variation in field populations of baculoviruses: Mechanisms for generating variation and its potential role in baculovirus epizootiology. Virol. Sin. 2009, 24, 458–469. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Pruijssers, A.J.; Vlak, J.M. Identification of pif-2, a third conserved baculovirus gene required for per os infection of insects. J. Gen. Virol. 2003, 84, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Serrano, A.; Pijlman, G.P.; Vlak, J.M.; Muñoz, D.; Williams, T.; Caballero, P. Identification of Spodoptera exigua nucleopolyhedrovirus genes involved in pathogenicity and virulence. J. Invertebr. Pathol. 2015, 126, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Moscardi, F. Use of viruses for pest control in Brazil: The case of the nuclear polyhedrosis virus of the soybean caterpillar, Anticarsia gemmatalis. Mem. Inst. Oswaldo Cruz 1989, 84, 51–56. [Google Scholar] [CrossRef]
- Moscardi, F. Assessment of the application of baculoviruses for control of Lepidoptera. Annu. Rev. Entomol. 1999, 44, 257–289. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Maruniak, A.; Pavan, O.H.; Maruniak, J.E. A variable region of Anticarsia gemmatalis nuclear polyhedrosis virus contains tandemly repeated DNA sequences. Virus Res. 1996, 41, 123–132. [Google Scholar] [CrossRef]
- Batista, T.F.C. Fatores Que Limitam a Eficiencia de Baculovirus Anticarsia Sobre Anticarsia gemmatalis Hubner, 1818; UFPEL: Pelotas, Brazil, 1997. [Google Scholar]
- Berino, E. Determinação da Atividade Biológica de Isolados Geográficos e Temporais de VPN de Anticarsia gemmatalis Hübner, 1818 (Lep., Noctuidae). Master’s Thesis, Universidade Federal do Rio Grande do Sul, Porto Alegre, Brazil, 1995. [Google Scholar]
- Cory, J.S.; Myers, J.H. The ecology and evolution of insect baculoviruses. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 239–272. [Google Scholar] [CrossRef]
- Sović, I.; Križanović, K.; Skala, K.; Šikić, M. Evaluation of hybrid and non-hybrid methods for de novo assembly of nanopore reads. Bioinformatics 2016, 32, 2582–2589. [Google Scholar] [CrossRef] [PubMed]
- Asser-Kaiser, S.; Fritsch, E.; Undorf-Spahn, K.; Kienzle, J.; Eberle, K.E.; Gund, N.A.; Reineke, A.; Zebitz, C.P.W.; Heckel, D.G.; Huber, J.; et al. Rapid Emergence of Baculovirus Resistance in Codling Moth Due to Dominant, Sex-Linked Inheritance. Science 2007, 317, 1916–1918. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.D. The Development and Evaluation of Cryptophlebia Leucotreta Granulovirus (CrleGV) as a Biological Control Agent for the Management of False Codling Moth, Cryptophlebia Leucotreta, on Citrus; Rhodes University: Grahamstown, South Africa, 2002. [Google Scholar]
- Moore, S.D.; Hendry, D.A.; Richards, G.I. Virulence of a South African isolate of the Cryptophlebia leucotreta granulovirus to Thaumatotibia leucotreta neonate larvae. BioControl 2011, 56, 341–352. [Google Scholar] [CrossRef]
- Hunter-Fujita, F.R.; Entwistle, P.F.; Evans, H.F.; Crook, N.E. Insect Viruses and Pest Management; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 1998. [Google Scholar]
- Loman, N.J.; Quinlan, A.R. Poretools: A toolkit for analyzing nanopore sequence data. Bioinformatics 2014, 30, 3399–3401. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef] [PubMed]
- Finney, D.J. Probit Analysis, 3rd ed.; Cambridge University Press: Cambridge, UK, 1971. [Google Scholar]
- Van Ark, H. Introduction to the analysis of quantal response. Agric. Res. Counc. Agrimetrics Inst. Pretoria 1995, 78, 13–17. [Google Scholar]
- Abbott, W.S. A method of computing the effectiveness of an insecticide. J. Econ. Entomol. 1925, 18, 265–267. [Google Scholar] [CrossRef]

| Nucleotide Position | 21022 | 21222 | 22400 | 22404 | 22414 | 22418 | 22427 | 22431 | 22445 | 38193 | 38194 | 38195 | 39138 | 39139 | 39140 | 39141 | 39142 | 39143 | 39144 | 39145 | 39146 | 39147 | 39148 | 39149 | 39150 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | 5′ | 45 | 3′ | 3′ | 46 | 5′ | ||||||||||||||||||||
| Gene & Frame | - | - | - | - | - | - | - | - | - | PIF2 | HP | |||||||||||||||
| 2015 | NT | T | G | A | A | A | A | A | A | A | A | T | T | G | T | T | A | A | A | A | T | T | - | - | - | - |
| AA | I | N | F | N | ||||||||||||||||||||||
| 2012 | NT | Y | C | A | A | A | A | A | A | A | A | C | T | G | T | T | A | A | A | A | T | T | - | - | - | - |
| AA | T | N | F | N | ||||||||||||||||||||||
| 2009 | NT | C | C | A | A | A | A | A | A | A | A | T | T | G | T | T | A | A | A | A | T | T | - | - | - | - |
| AA | I | N | F | N | ||||||||||||||||||||||
| 2007 | NT | Y | C | A | A | A | A | A | A | A | A | T | T | G | T | T | A | A | A | A | T | T | C | G | C | G |
| AA | I | * | F | E | R | |||||||||||||||||||||
| 2005 | NT | T | C | T | T | T | T | T | T | T | A | T | T | G | T | T | A | A | A | A | T | T | C | G | C | G |
| AA | I | * | F | E | R | |||||||||||||||||||||
| 2003 | NT | C | C | W | W | W | W | W | W | W | A | T | T | G | T | T | A | A | A | A | T | T | - | - | - | - |
| AA | I | N | F | N | ||||||||||||||||||||||
| 2000 | NT | C | C | W | W | W | W | W | W | W | A | T | T | G | T | T | A | A | A | A | T | T | - | - | - | - |
| AA | I | N | F | N | ||||||||||||||||||||||
| Nucleotide Position | 62164 | 62165 | 62166 | 62167 | 62168 | 62169 | 62170 | 62171 | 62172 | 62173 | 62174 | 62175 | 62176 | 62177 | 62178 | 62179 | 62180 | 62181 | 62182 | 62183 | 62184 | 62185 | 62186 | 62187 | 62188 | 62189 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | 5′ | 75 | 3′ | 5′ | 75 | 3′ | |||||||||||||||||||||
| Gene & Frame | HP | HP | |||||||||||||||||||||||||
| 2015 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2012 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2009 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2007 | NT | T | A | C | A | T | A | T | C | G | T | A | G | G | T | T | A | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2005 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2003 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | Y | I | S | * | |||||||||||||||||||||||
| 2000 | NT | T | A | - | - | - | - | - | - | - | - | - | - | - | - | - | - | T | A | T | A | T | C | G | T | A | G |
| AA | I | S | * | ||||||||||||||||||||||||
| Nucleotide Position | 70584 | 70585 | 70586 | 75271 | 75272 | 79953 | 79954 | 79955 | 94105 | 94106 | 94107 | 101133 | 101134 | 101135 | 104415 | 104416 | 104417 | 104593 | 104594 | 104595 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF | 5′ | 81 | 3′ | 5′ | 94 | 3′ | 5′ | 109 | 3′ | 3′ | 117 | 5′ | 3′ | 119 | 5′ | 5′ | 120 | 3′ | |||
| Gene & Frame | P33 | - | - | AC81 | HP | HP | LEF8 | HP | |||||||||||||
| 2015 | NT | G | A | C | - | - | T | T | A | A | G | A | A | G | T | G | T | T | A | T | G |
| AA | D | L | R | T | N | M | |||||||||||||||
| 2012 | NT | G | A | C | - | - | T | T | G | A | G | A | A | G | T | A | T | T | A | C | G |
| AA | D | L | R | T | N | T | |||||||||||||||
| 2009 | NT | G | A | C | - | - | T | T | R | A | G | A | A | R | T | R | T | T | A | Y | G |
| AA | D | L | R | I/T | N | T/M | |||||||||||||||
| 2007 | NT | G | A | T | - | - | T | T | R | A | G | A | A | A | T | G | T | T | A | C | G |
| AA | D | L | R | I | N | - | |||||||||||||||
| 2005 | NT | G | A | T | C | A | T | T | G | A | G | A | A | A | T | G | T | T | A | C | G |
| AA | D | L | R | I | N | - | |||||||||||||||
| 2003 | NT | G | A | T | - | - | T | T | G | A | A | A | A | A | T | G | T | T | A | C | G |
| AA | D | L | K | I | N | - | |||||||||||||||
| 2000 | NT | G | A | T | - | - | T | T | G | A | A | A | A | A | T | G | T | T | A | C | G |
| AA | D | L | K | I | N | - | |||||||||||||||
| Sample | Lethal Concentration | Standard Error (SE) | 95% Fiducial Limits | Means of Empirical Probits | Slope ± SE | ||
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| 2000 | LC50 | 4.071 × 103 | ± 6.286 × 102 | 2.974 × 103 | 5.496 × 103 | 5.1073 | 0.9340 ± 0.0765 |
| LC90 | 9.590 × 104 | ± 2.702 × 104 | 5.882 × 104 | 1.822 × 105 | |||
| 2015 | LC50 | 1.170 × 103 | ± 2.072 × 102 | 8.095 × 102 | 1.636 × 103 | 5.2293 | 0.7527 ± 0.0608 |
| LC90 | 7.849 × 104 | ± 1.826 × 104 | 3.455 × 104 | 1.193 × 105 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van der Merwe, M.; Jukes, M.D.; Rabalski, L.; Knox, C.; Opoku-Debrah, J.K.; Moore, S.D.; Krejmer-Rabalska, M.; Szewczyk, B.; Hill, M.P. Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide. Int. J. Mol. Sci. 2017, 18, 2327. https://doi.org/10.3390/ijms18112327
Van der Merwe M, Jukes MD, Rabalski L, Knox C, Opoku-Debrah JK, Moore SD, Krejmer-Rabalska M, Szewczyk B, Hill MP. Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide. International Journal of Molecular Sciences. 2017; 18(11):2327. https://doi.org/10.3390/ijms18112327
Chicago/Turabian StyleVan der Merwe, Marcel, Michael D. Jukes, Lukasz Rabalski, Caroline Knox, John K. Opoku-Debrah, Sean D. Moore, Martyna Krejmer-Rabalska, Boguslaw Szewczyk, and Martin P. Hill. 2017. "Genome Analysis and Genetic Stability of the Cryptophlebia leucotreta Granulovirus (CrleGV-SA) after 15 Years of Commercial Use as a Biopesticide" International Journal of Molecular Sciences 18, no. 11: 2327. https://doi.org/10.3390/ijms18112327