Comparative Proteomic Analysis of Lysine Acetylation in Fish CIK Cells Infected with Aquareovirus
Abstract
:1. Introduction
2. Results
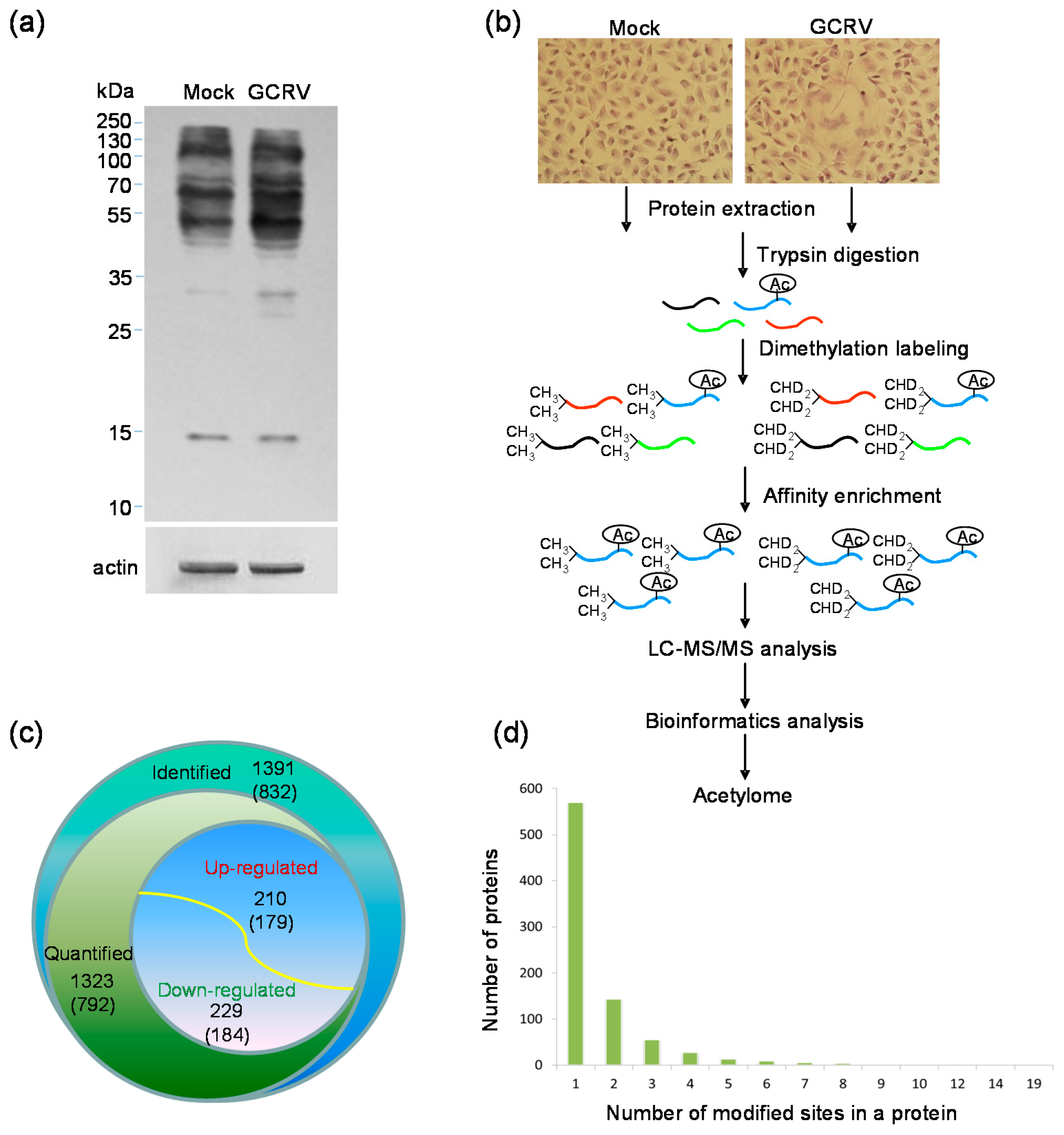
2.1. Quantitative Analysis of Lysine Acetylation in CIK Cells in Response to GCRV Infection
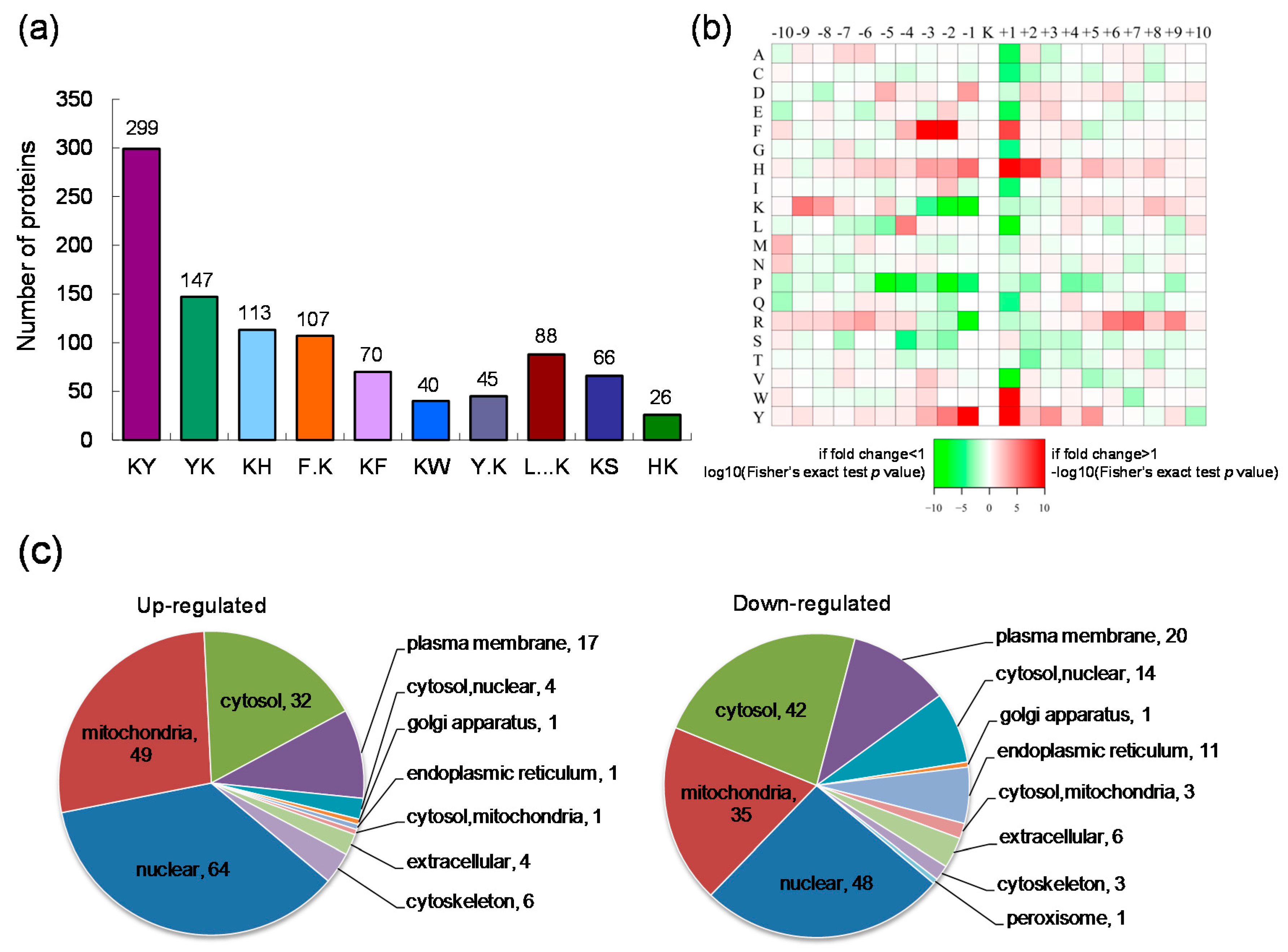
2.2. Motif Analysis of the Identified Lysine Acetylation Sites and Subcellular Localization of the Differentially Expressed Lysine Acetylated Proteins
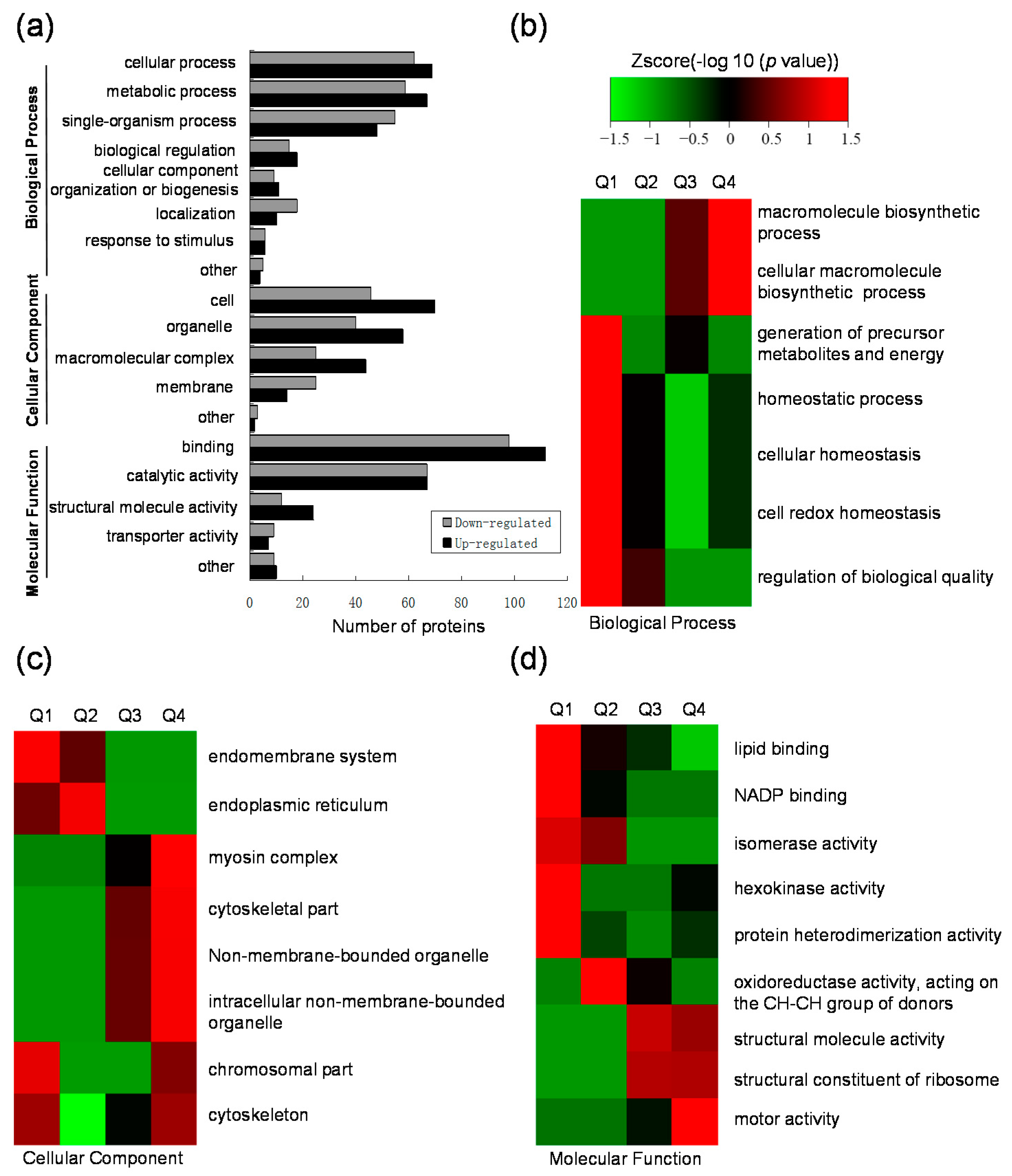
2.3. Gene Ontology (GO) Functional Classification and GO Enrichment-Based Clustering Analysis of the Differentially Expressed Lysine Acetylated Proteins
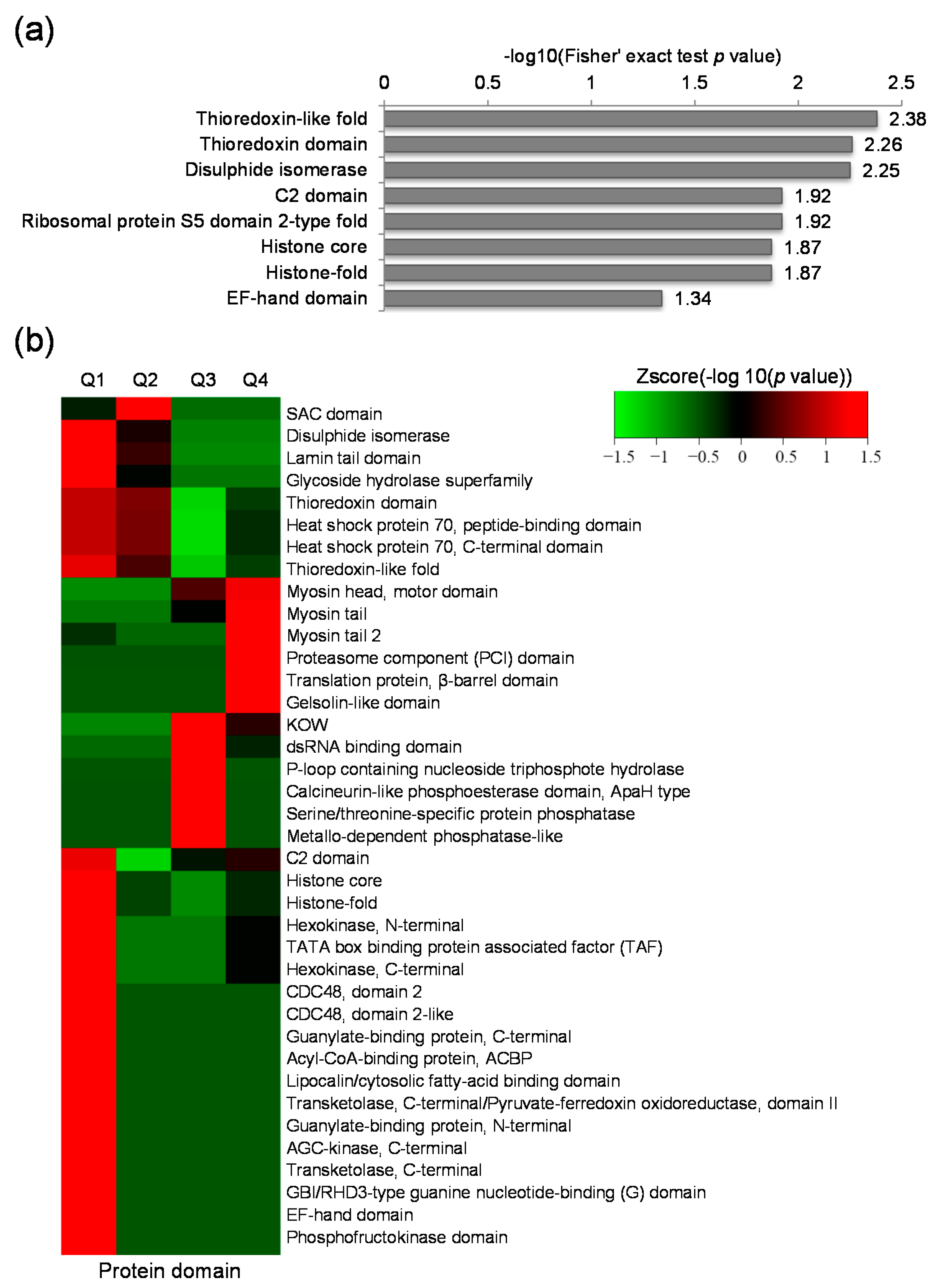
2.4. Protein Domain Based Enrichment and Clustering Analysis of the Differentially Expressed Lysine Acetylated Proteins
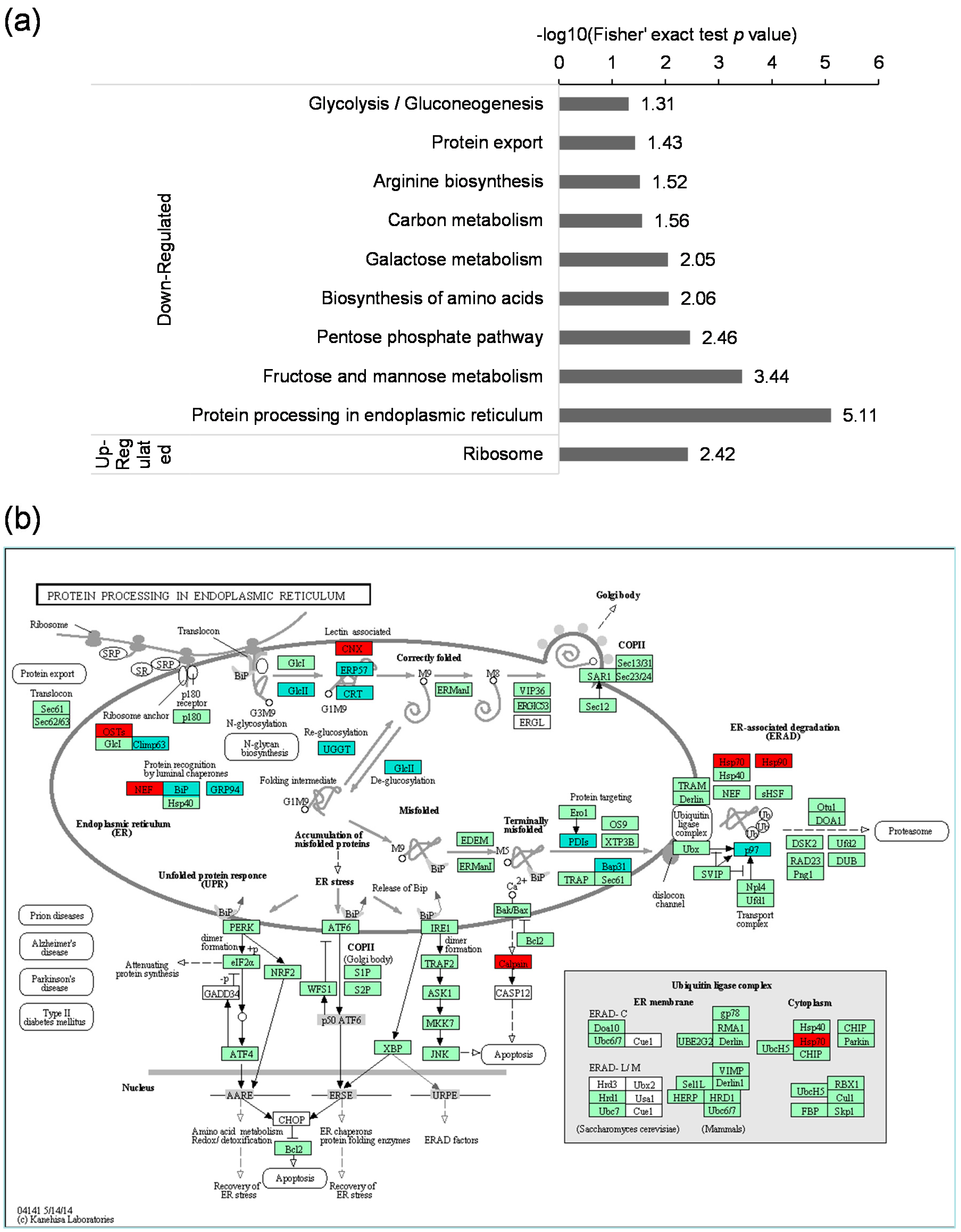
2.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis of the Differentially Expressed Lysine Acetylated Proteins
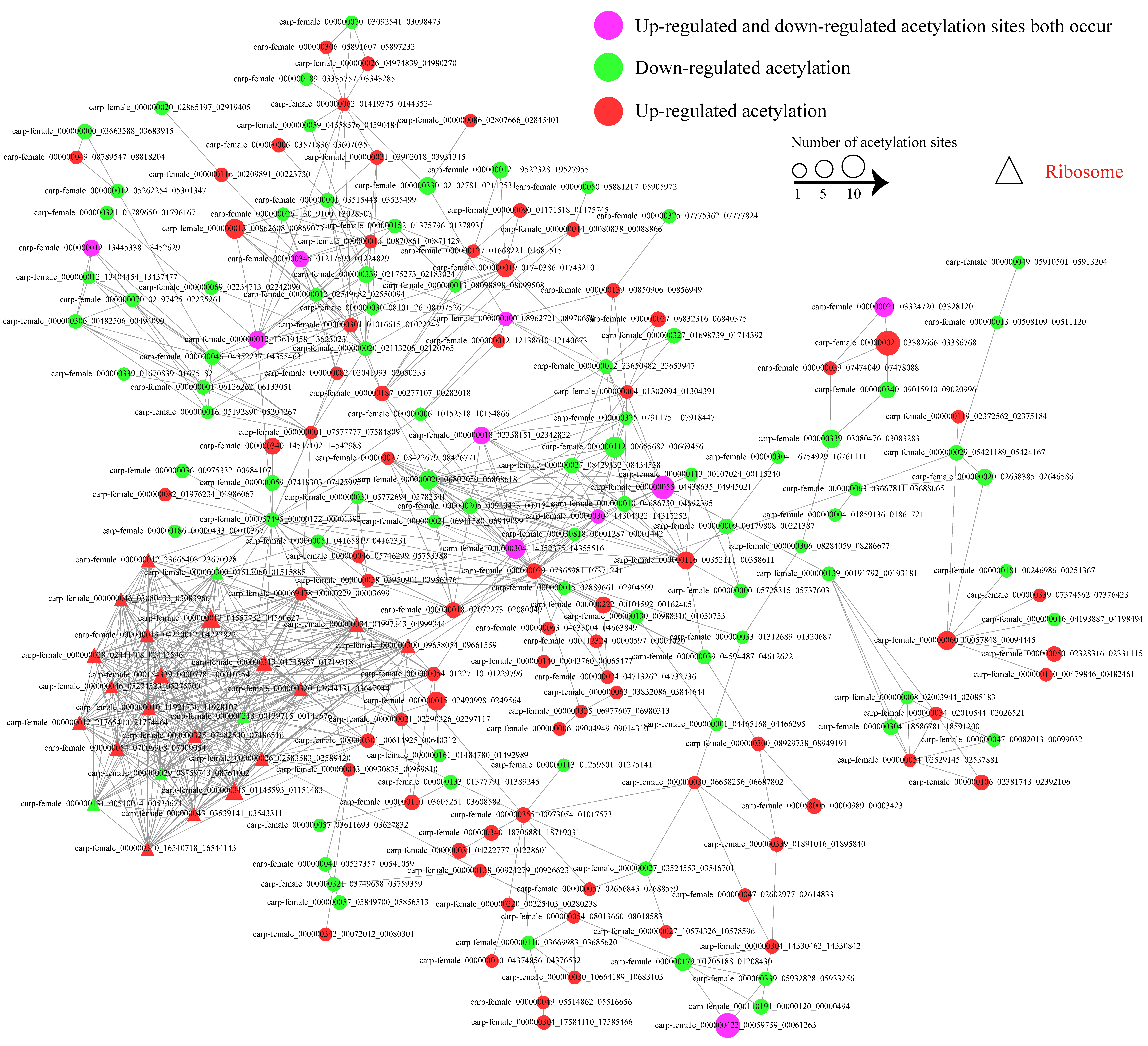
2.6. Protein–Protein Interaction (PPI) Network Analysis of Differentially Expressed Proteins
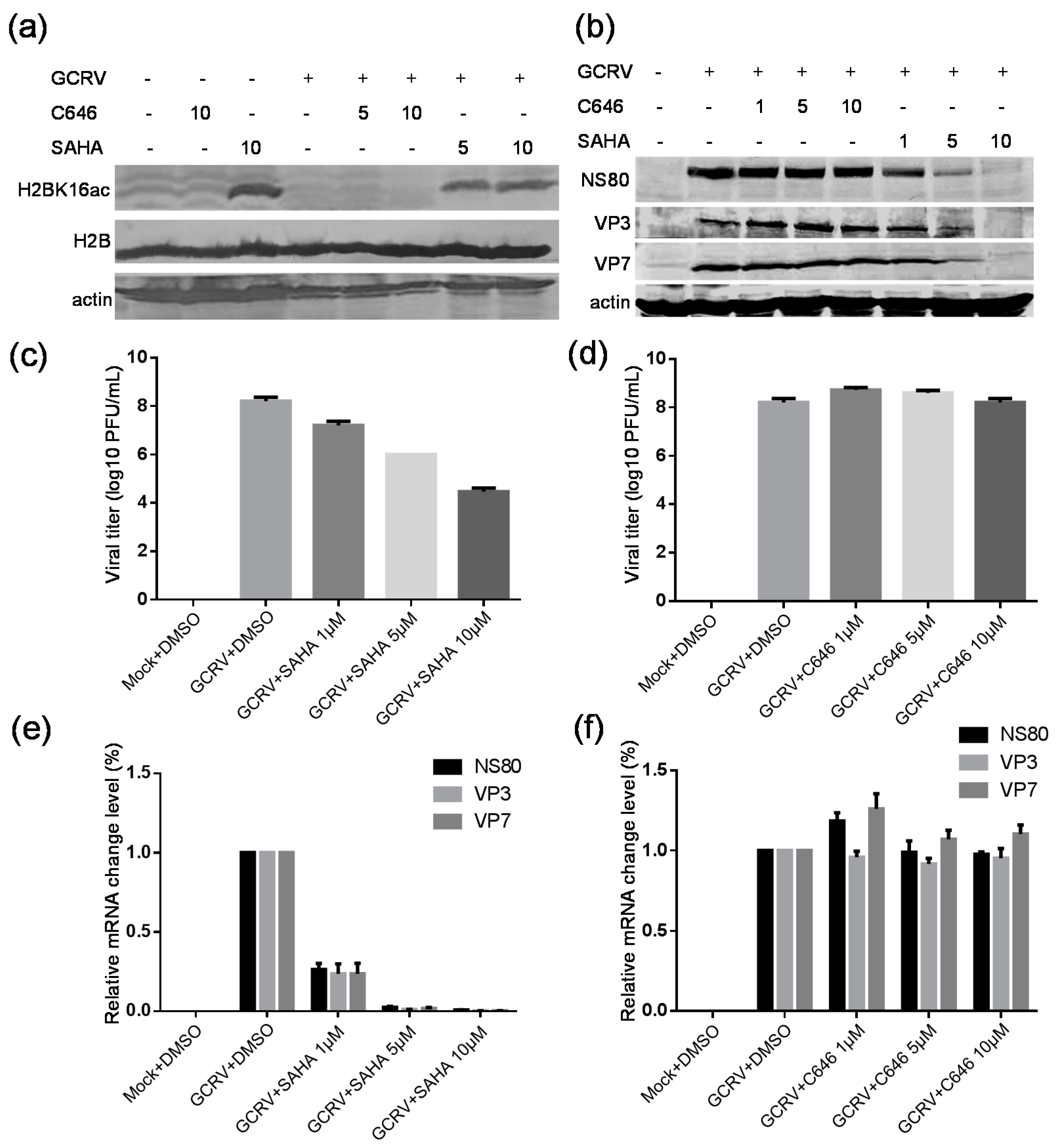
2.7. Suberoylanilide Hydroxamic Acid (SAHA) Treatment Inhibited the GCRV Infection, Whereas C646 Treatment Had No Obvious Effect on Virus Replication
3. Discussion
4. Materials and Methods
4.1. Cell, Virus and Antibodies
4.2. Viral Infection
4.3. Protein Extraction and Western Blotting Analysis
4.4. Trypsin Digestion, Dimethylation Labeling and Affinity Enrichment
4.5. LC–MS/MS Analysis
4.6. Database Search
4.7. Bioinformatics Analysis
4.8. Pharmacological Inhibitors and Cell Viability Assay
4.9. Analysis of Viral Growth after Treatment with Drugs
4.10. Real-Time Quantitative PCR Analysis
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GCRV | Grass carp reovirus |
| CIK | Ctenopharyngodon idella kidney |
| HDAC | Histone deacetylase |
| SAHA | Suberoylanilide hydroxamic acid |
| PTM | Post-translational modification |
| HAT | Histone acetyltransferase |
| HIV | Human immunodeficiency virus |
| HPV | Human papilloma virus |
| RSV | Respiratory syncytial virus |
| HCV | Hepatitis C virus |
| KRT | Keratin |
| PDIA3 | Protein disulfide isomerase family A member 3 |
| GO | Gene ontology |
| PCI domain | Proteasome component domain |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PPI | Protein–protein interaction |
| STRING | Search tool for the retrieval of interacting genes |
| MOI | Multiplicity of infection |
| Hsps | Heat shock proteins |
| eEF | Eukaryotic elongation factor |
| canx | Calnexin |
| MEM | Minimal essential medium |
| FBS | Fetal bovine serum |
| h.p.i. | Hours post-infection |
| PVDF | Polyvinylidene fluoride |
| TBST | Tris-buffered saline-tween |
References
- King, A.M.Q.; Adams, M.J.; Cartens, E.B.; Lefkowitz, E.J. Virus Taxonomy, Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: San Diego, CA, USA, 2011. [Google Scholar]
- Ahne, W. Viral infections of aquatic animals with special reference to Asian aquaculture. Annu. Rev. Fish Dis. 1994, 4, 375–388. [Google Scholar] [CrossRef]
- Fang, Q.; Ke, L.H.; Cai, Y.Q. Growth characterization and high titer culture of GCHV. Virol. Sin. 1989, 4, 315–319. [Google Scholar]
- Ke, L.H.; Yu, L.F.; Fang, Q.; Ding, Q.Q.; Cai, Y.Q. Characteristics of a novel isolate of grass carp hemorrhage virus (GCHV). Acta Hydrobiol. Sin. 1990, 14, 153–159. [Google Scholar]
- Zhang, X.; Jin, L.; Fang, Q.; Hui, W.H.; Zhou, Z.H. 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 2010, 141, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Sun, X.; Yan, L.; Shao, L.; Fang, Q. The NS16 protein of aquareovirus-C is a fusion-associated small transmembrane (FAST) protein, and its activity can be enhanced by the nonstructural protein NS26. Virus Res. 2013, 171, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, H.; Chen, Q.; Zhang, F.; Fang, Q. The N-Terminal of Aquareovirus NS80 Is Required for Interacting with Viral Proteins and Viral Replication. PLoS ONE 2016, 11, e0148550. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhang, J.; Guo, H.; Yan, L.; Chen, Q.; Zhang, F.; Fang, Q. VP5 autocleavage is required for efficient infection by in vitro-recoated aquareovirus particles. J. Gen. Virol. 2015, 96, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Rangel, A.A.; Rockemann, D.D.; Hetrick, F.M.; Samal, S.K. Identification of grass carp haemorrhage virus as a new genogroup of aquareovirus. J. Gen. Virol. 1999, 80 Pt 9, 2399–2402. [Google Scholar] [CrossRef] [PubMed]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed]
- Glozak, M.A.; Sengupta, N.; Zhang, X.; Seto, E. Acetylation and deacetylation of non-histone proteins. Gene 2005, 363, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Venne, A.S.; Kollipara, L.; Zahedi, R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics 2014, 14, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.W.; Yang, X.J. Comprehensive lysine acetylomes emerging from bacteria to humans. Trends Biochem. Sci. 2011, 36, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Yoon, E.K.; Chung, H.J.; Park, S.Y.; Hong, K.M.; Lee, C.H.; Lee, Y.S.; Choi, K.; Yang, Y.; Kim, K.; et al. p53 acetylation enhances Taxol-induced apoptosis in human cancer cells. Apoptosis 2013, 18, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Birks, S.M.; Danquah, J.O.; King, L.; Vlasak, R.; Gorecki, D.C.; Pilkington, G.J. Targeting the GD3 acetylation pathway selectively induces apoptosis in glioblastoma. Neuro Oncol. 2011, 13, 950–960. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, W.; Jiang, W.; Yu, W.; Lin, Y.; Zhang, T.; Yao, J.; Zhou, L.; Zeng, Y.; Li, H.; et al. Regulation of cellular metabolism by protein lysine acetylation. Science 2010, 327, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Contreras, L.; Hernandez-Ramirez, V.I.; Lagunes-Guillen, A.E.; Montano, S.; Chavez-Munguia, B.; Sanchez-Ramirez, B.; Talamas-Rohana, P. Exploring the possible role of lysine acetylation on Entamoeba histolytica virulence: A focus on the dynamics of the actin cytoskeleton. Biomed. Res. Int. 2013, 2013, 757392. [Google Scholar] [CrossRef] [PubMed]
- Banreti, A.; Sass, M.; Graba, Y. The emerging role of acetylation in the regulation of autophagy. Autophagy 2013, 9, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, Y.; Eugene Chin, Y.; Xia, Q. The role of acetylation in TLR4-mediated innate immune responses. Immunol. Cell Biol. 2013, 91, 611–614. [Google Scholar] [CrossRef] [PubMed]
- Herbein, G.; Wendling, D. Histone deacetylases in viral infections. Clin. Epigenet. 2010, 1, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Su, Z.; Song, S.; Chiu, H.; Zhang, B.; Yi, L.; Tian, M.; Wang, H. Histone deacetylase inhibitors suppress RSV infection and alleviate virus-induced airway inflammation. Int. J. Mol. Med. 2016, 38, 812–822. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Saito, Y.; Sugiyama, K.; Sakasegawa, N.; Muramatsu, T.; Fukuda, S.; Yoneya, M.; Kimura, M.; Ebinuma, H.; Hibi, T.; et al. Suppressive effect of the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) on hepatitis C virus replication. J. Cell. Biochem. 2013, 114, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Fukuyama, S.; Sakai-Tagawa, Y.; Takashita, E.; Shoemaker, J.E.; Kawaoka, Y. C646, a Novel p300/CREB-Binding Protein-Specific Inhibitor of Histone Acetyltransferase, Attenuates Influenza A Virus Infection. Antimicrob. Agents Chemother. 2015, 60, 1902–1906. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, S.; Bode, L.; Liu, C.; Zhang, L.; Wang, X.; Li, D.; Lei, Y.; Peng, X.; Cheng, Z.; et al. Persistent human Borna disease virus infection modifies the acetylome of human oligodendroglia cells towards higher energy and transporter levels. Virology 2015, 485, 58–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, L.; Yang, Y.; Bode, L.; Huang, H.; Liu, C.; Huang, R.; Zhang, L.; Wang, X.; Zhang, L.; et al. Human borna disease virus infection impacts host proteome and histone lysine acetylation in human oligodendroglia cells. Virology 2014, 464–465, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.K.; Kim, S.; Lee, S. Global proteomic analysis of lysine acetylation in zebrafish (Danio rerio) embryos. Electrophoresis 2016, 37, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.K.; Sim, J.; Kim, S.J.; Oh, H.R.; Nam, D.H.; Lee, S. Global proteomic analysis of protein acetylation affecting metabolic regulation in Daphnia pulex. Biochimie 2016, 121, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Liu, P.; Wang, J.; Wang, Y.; Hou, L.; Gu, W.; Wang, W. Systematic analysis of the lysine acetylome of the pathogenic bacterium Spiroplasma eriocheiris reveals acetylated proteins related to metabolism and helical structure. J. Proteom. 2016, 148, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, W.; Cao, L.; Li, X.; He, T.; Wu, Z.; Li, W. SAHA treatment reveals the link between histone lysine acetylation and proteome in nonsmall cell lung cancer A549 Cells. J. Proteome Res. 2013, 12, 4064–4073. [Google Scholar] [CrossRef] [PubMed]
- Bowers, E.M.; Yan, G.; Mukherjee, C.; Orry, A.; Wang, L.; Holbert, M.A.; Crump, N.T.; Hazzalin, C.A.; Liszczak, G.; Yuan, H.; et al. Virtual ligand screening of the p300/CBP histone acetyltransferase: Identification of a selective small molecule inhibitor. Chem. Biol 2010, 17, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Makino, T.; Yamasaki, M.; Takeno, A.; Shirakawa, M.; Miyata, H.; Takiguchi, S.; Nakajima, K.; Fujiwara, Y.; Nishida, T.; Matsuura, N.; et al. Cytokeratins 18 and 8 are poor prognostic markers in patients with squamous cell carcinoma of the oesophagus. Br. J. Cancer 2009, 101, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Wang, H.; Liu, Y.; Ding, F.; Ni, Y.; Shao, S. High KRT8 expression promotes tumor progression and metastasis of gastric cancer. Cancer Sci. 2017, 108, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Wiche, G.; Osmanagic-Myers, S.; Castanon, M.J. Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 2015, 32, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Moch, M.; Neckernuss, T.; Paschke, S.; Herrmann, H.; Marti, O. Both monovalent cations and plectin are potent modulators of mechanical properties of keratin K8/K18 networks. Soft Matter 2016, 12, 6964–6974. [Google Scholar] [CrossRef] [PubMed]
- Turano, C.; Gaucci, E.; Grillo, C.; Chichiarelli, S. ERp57/GRP58: A protein with multiple functions. Cell. Mol. Biol. Lett. 2011, 16, 539–563. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; Yang, Q.; Li, D.; Liang, W.; Song, L. Proteome-wide analysis of lysine acetylation in the plant pathogen Botrytis cinerea. Sci. Rep. 2016, 6, 29313. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Wang, X.; Zeng, J.; Zhou, M.; Duan, X.; Li, Q.; Zhang, Z.; Luo, H.; Pang, L.; Li, W.; et al. Proteome-wide lysine acetylation profiling of the human pathogen Mycobacterium tuberculosis. Int. J. Biochem. Cell Biol. 2015, 59, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Zhu, H.; Zhou, Y.; Wu, C.; Liu, Y.; Sheng, Q.; Lv, Z.; Zhang, W.; Yu, W.; Jiang, C.; et al. Comprehensive profiling of lysine acetylation suggests the widespread function is regulated by protein acetylation in the silkworm, Bombyx mori. Proteomics 2015, 15, 3253–3266. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Wang, C.; Han, Y.; Liu, Z.; Wu, T.; Liu, Y.; Liu, Y.; Tan, Y.; Cai, X.; Cao, Y.; et al. Identification of Lysine Acetylation in Mycobacterium abscessus Using LC-MS/MS after Immunoprecipitation. J. Proteome Res. 2016, 15, 2567–2578. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sprung, R.; Pei, J.; Tan, X.; Kim, S.; Zhu, H.; Liu, C.F.; Grishin, N.V.; Zhao, Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteom. 2009, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Lundby, A.; Lage, K.; Weinert, B.T.; Bekker-Jensen, D.B.; Secher, A.; Skovgaard, T.; Kelstrup, C.D.; Dmytriyev, A.; Choudhary, C.; Lundby, C.; et al. Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2012, 2, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sellers, J.R. Myosins: A diverse superfamily. Biochim. Biophys. Acta 2000, 1496, 3–22. [Google Scholar] [CrossRef]
- Rayment, I.; Holden, H.M.; Whittaker, M.; Yohn, C.B.; Lorenz, M.; Holmes, K.C.; Milligan, R.A. Structure of the actin-myosin complex and its implications for muscle contraction. Science 1993, 261, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Ghoshdastider, U.; Popp, D.; Burtnick, L.D.; Robinson, R.C. The expanding superfamily of gelsolin homology domain proteins. Cytoskeleton 2013, 70, 775–795. [Google Scholar] [CrossRef] [PubMed]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P.; Le Breton, L. Hsp90: Breaking the symmetry. Mol. Cell 2015, 58, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Tripal, P.; Bauer, M.; Naschberger, E.; Mortinger, T.; Hohenadl, C.; Cornali, E.; Thurau, M.; Sturzl, M. Unique features of different members of the human guanylate-binding protein family. J. Interferon Cytokine Res. 2007, 27, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Baek, G.H.; Cheng, H.; Choe, V.; Bao, X.; Shao, J.; Luo, S.; Rao, H. Cdc48: A swiss army knife of cell biology. J. Amino Acids 2013, 2013, 183421. [Google Scholar] [CrossRef] [PubMed]
- Desmet, E.A.; Anguish, L.J.; Parker, J.S. Virus-mediated compartmentalization of the host translational machinery. MBio 2014, 5, e01463-14. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef] [PubMed]
- Wallin, R.P.; Lundqvist, A.; More, S.H.; von Bonin, A.; Kiessling, R.; Ljunggren, H.G. Heat-shock proteins as activators of the innate immune system. Trends Immunol. 2002, 23, 130–135. [Google Scholar] [CrossRef]
- Riis, B.; Rattan, S.I.; Clark, B.F.; Merrick, W.C. Eukaryotic protein elongation factors. Trends Biochem. Sci. 1990, 15, 420–424. [Google Scholar] [CrossRef]
- Proud, C.G. Peptide-chain elongation in eukaryotes. Mol. Biol. Rep. 1994, 19, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Chevet, E.; Smirle, J.; Cameron, P.H.; Thomas, D.Y.; Bergeron, J.J. Calnexin phosphorylation: Linking cytoplasmic signalling to endoplasmic reticulum lumenal functions. Semin. Cell Dev. Biol. 2010, 21, 486–490. [Google Scholar] [CrossRef] [PubMed]
- Delom, F.; Emadali, A.; Cocolakis, E.; Lebrun, J.J.; Nantel, A.; Chevet, E. Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ. 2007, 14, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Weinert, B.T.; Nishida, Y.; Verdin, E.; Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 2014, 15, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.M.; Voss, A.K.; Thomas, T.; Allan, R.S. Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J. Leukoc. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dorfel, M.J.; Lyon, G.J. The biological functions of Naa10—From amino-terminal acetylation to human disease. Gene 2015, 567, 103–131. [Google Scholar] [CrossRef] [PubMed]
- Ree, R.; Myklebust, L.M.; Thiel, P.; Foyn, H.; Fladmark, K.E.; Arnesen, T. The N-terminal acetyltransferase Naa10 is essential for zebrafish development. Biosci. Rep. 2015, 35. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, G.A.; Zhang, K.; McBrian, M.A.; Grunstein, M.; Kurdistani, S.K.; Berk, A.J. Adenovirus small e1a alters global patterns of histone modification. Science 2008, 321, 1084–1085. [Google Scholar] [CrossRef] [PubMed]
- Suberbielle, E.; Stella, A.; Pont, F.; Monnet, C.; Mouton, E.; Lamouroux, L.; Monsarrat, B.; Gonzalez-Dunia, D. Proteomic analysis reveals selective impediment of neuronal remodeling upon Borna disease virus infection. J. Virol. 2008, 82, 12265–12279. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, J.; Guo, H.; Yan, S.; Chen, Q.; Zhang, F.; Fang, Q. Aquareovirus NS80 Initiates Efficient Viral Replication by Retaining Core Proteins within Replication-Associated Viral Inclusion Bodies. PLoS ONE 2015, 10, e0126127. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Seng, E.K.; Ding, Q.Q.; Zhang, L.L. Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch. Virol. 2008, 153, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Cheng, Z.; Zhu, J.; Xu, W.; Peng, X.; Chen, C.; Li, W.; Wang, F.; Cao, L.; Yi, X.; et al. Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci. Rep. 2015, 5, 9520. [Google Scholar] [CrossRef] [PubMed]
- Jin, W.; Wu, F. Proteome-Wide Identification of Lysine Succinylation in the Proteins of Tomato (Solanum lycopersicum). PLoS ONE 2016, 11, e0147586. [Google Scholar] [CrossRef] [PubMed]
| Histone and Modified Sites | Modified Sequence | Normalized Ratio H/L (↑↓) 1 |
|---|---|---|
| H1-βK168ac | K(ac)YPSVEMDK | 0.572 ↓ |
| H1.0K43ac | ATSHPK(ac)YSEMIK | 1.049 |
| H1.0K68ac | QSIQK(ac)YVK | 1.027 |
| H1.0K75ac | NHYK(ac)VGDNADSQIK | 1.272 |
| H2B3K16ac | K(ac)AVTK(ac)TQK | 2.215 ↑ |
| H2B3K20ac | K(ac)AVTK(ac)TQK | 2.215 ↑ |
| H2BK19ac | VTK(ac)TAGK(ac)SGK | 0.658 ↓ |
| H2BK23ac | VTK(ac)TAGK(ac)SGK | 0.678 |
| H2BK115ac | HAVSEGTK(ac)AVTK(ac)YTSSK | 0.789 |
| H2A.xK130ac | TGQAVASSGK(ac)SGK(ac)K | 0.307 ↓ |
| H2A.xK133ac | TGQAVASSGK(ac)SGK(ac)K | 1.232 |
| H4K19 | K(ac)QLATK(ac)AAR | 0.617 ↓ |
| H4K24 | QLATK(ac)AAR | 0.539 ↓ |
| H4K57 | YQK(ac)STELLIR | 0.89 |
| H4K80 | EIAQDFK(ac)TDLR | 0.808 |
| H4K123 | VTIMPK(ac)DIQLAR | 0.826 |
| MYST2K137 | CPTPGCNSLGHLTGK(ac)HER | 1.514 ↑ |
| Naa10K101 | FQISEVEPK(ac)YYADGEDAYAMK | 2.365 ↑ |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, H.; Zhang, J.; Wang, Y.; Bu, C.; Zhou, Y.; Fang, Q. Comparative Proteomic Analysis of Lysine Acetylation in Fish CIK Cells Infected with Aquareovirus. Int. J. Mol. Sci. 2017, 18, 2419. https://doi.org/10.3390/ijms18112419
Guo H, Zhang J, Wang Y, Bu C, Zhou Y, Fang Q. Comparative Proteomic Analysis of Lysine Acetylation in Fish CIK Cells Infected with Aquareovirus. International Journal of Molecular Sciences. 2017; 18(11):2419. https://doi.org/10.3390/ijms18112419
Chicago/Turabian StyleGuo, Hong, Jie Zhang, Yaping Wang, Chen Bu, Yanyan Zhou, and Qin Fang. 2017. "Comparative Proteomic Analysis of Lysine Acetylation in Fish CIK Cells Infected with Aquareovirus" International Journal of Molecular Sciences 18, no. 11: 2419. https://doi.org/10.3390/ijms18112419




