The Silencing of Carotenoid β-Hydroxylases by RNA Interference in Different Maize Genetic Backgrounds Increases the β-Carotene Content of the Endosperm
Abstract
:1. Introduction
2. Results
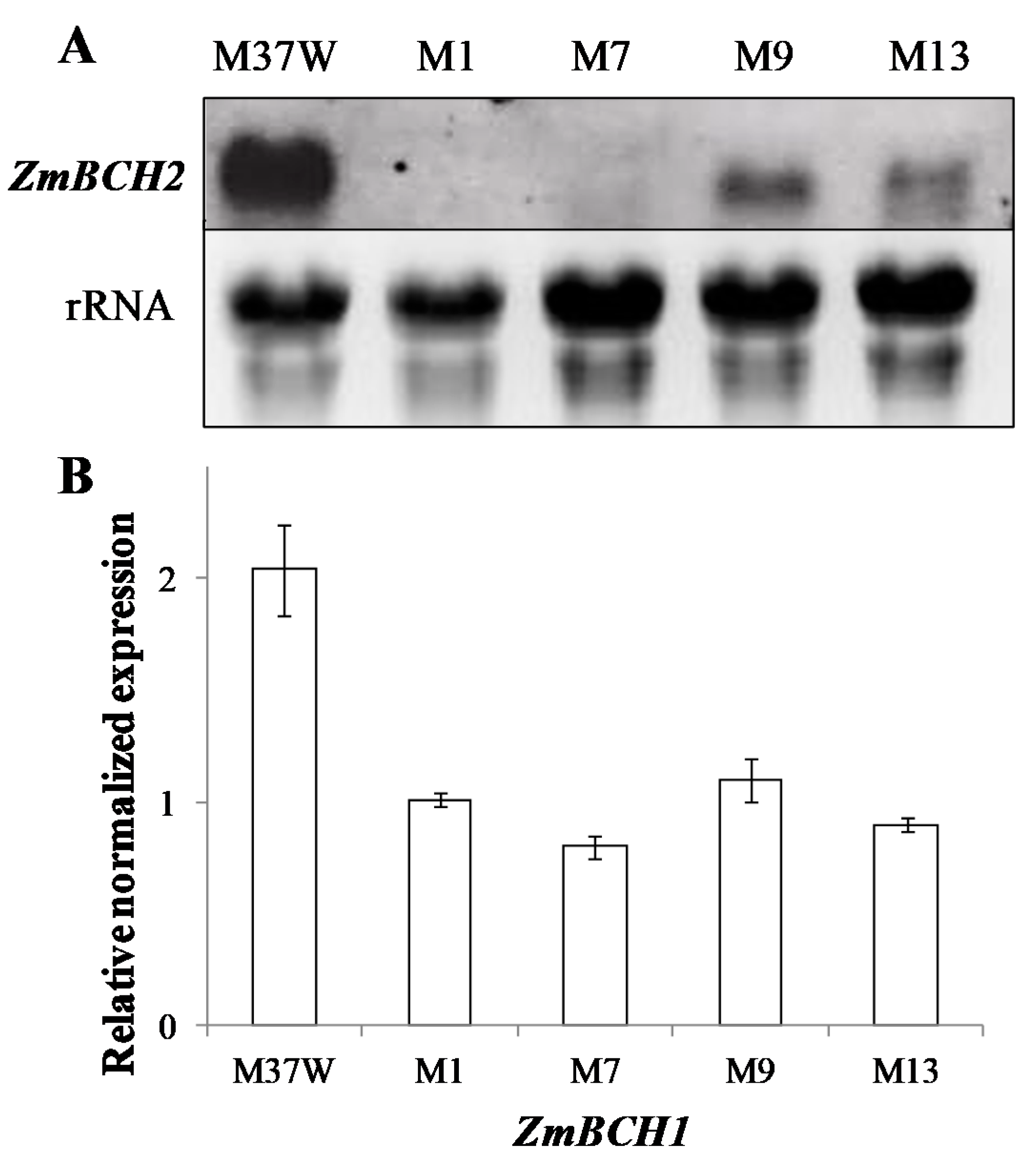
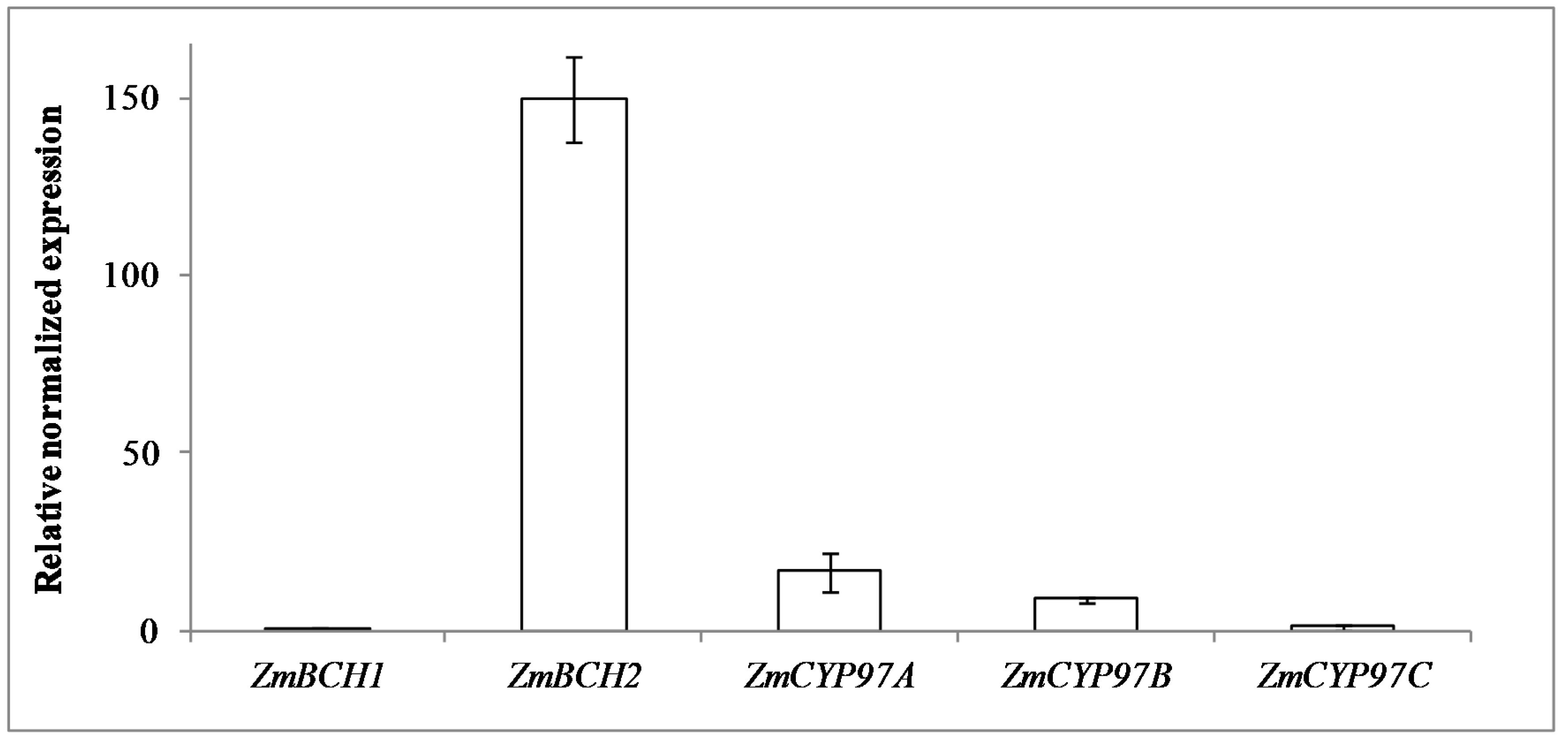
2.1. RNAi-Mediated Silencing of ZmBCH1 and ZmBCH2 in M37W White Maize
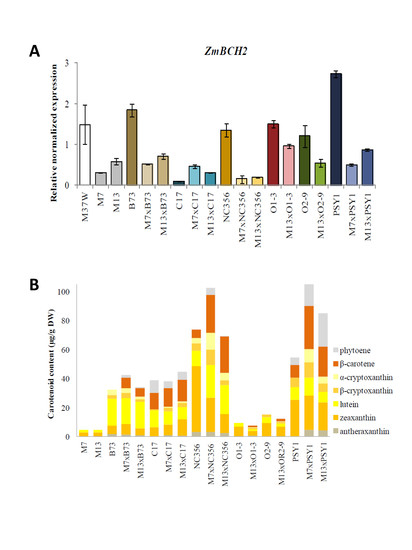
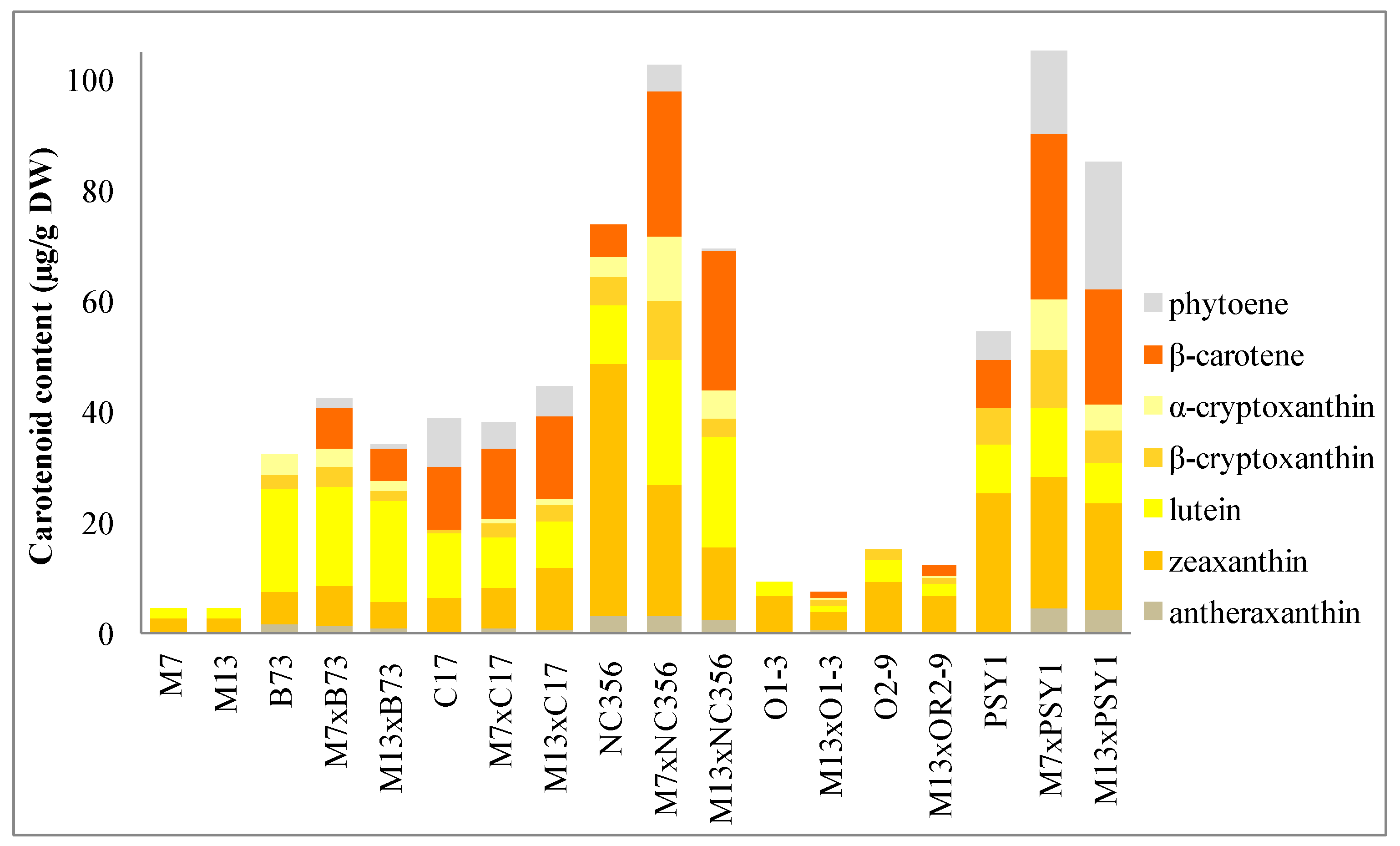
2.2. Carotenoid Profiles in Hybrids Derived from BCH-Silenced Parents
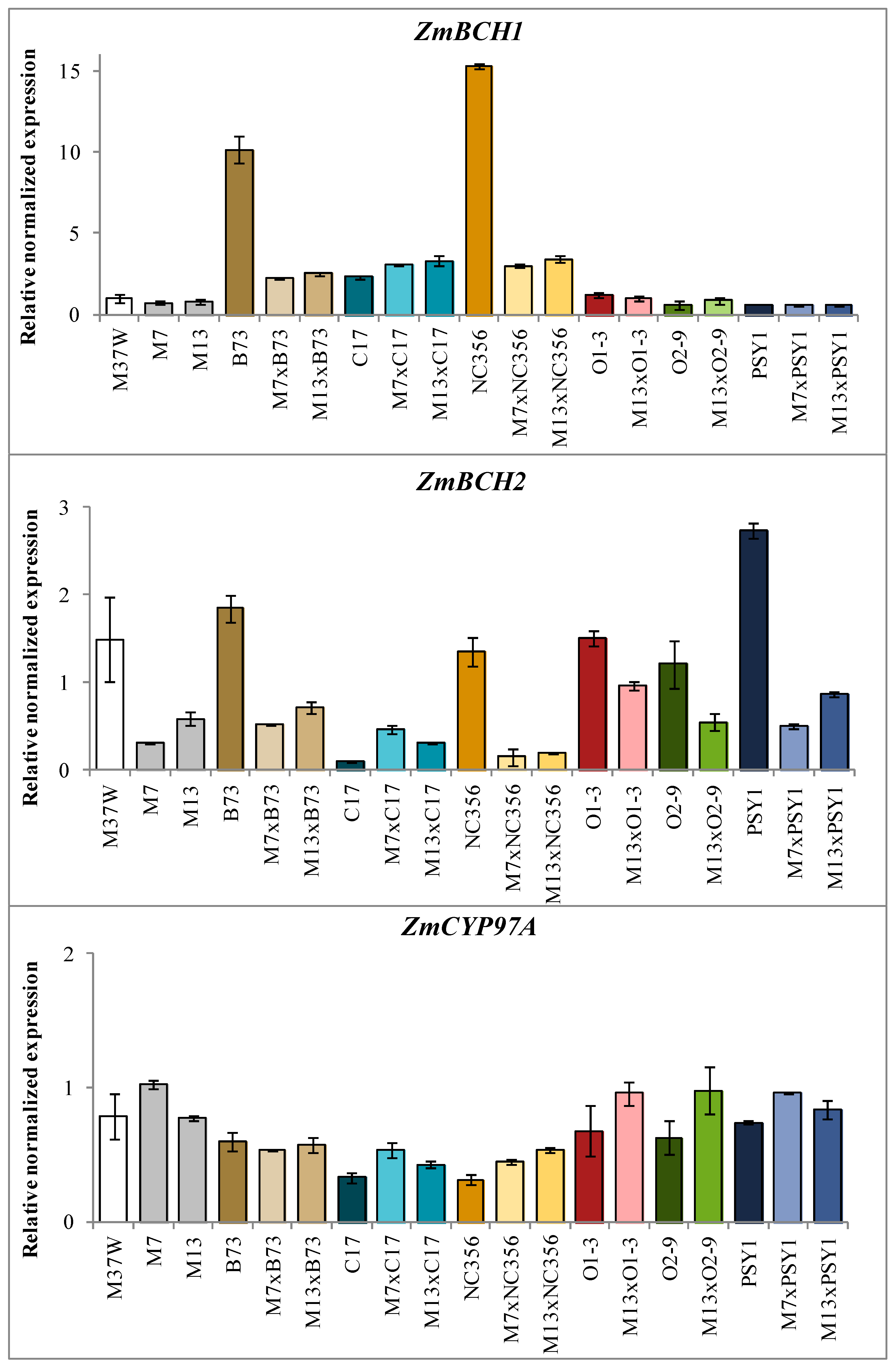
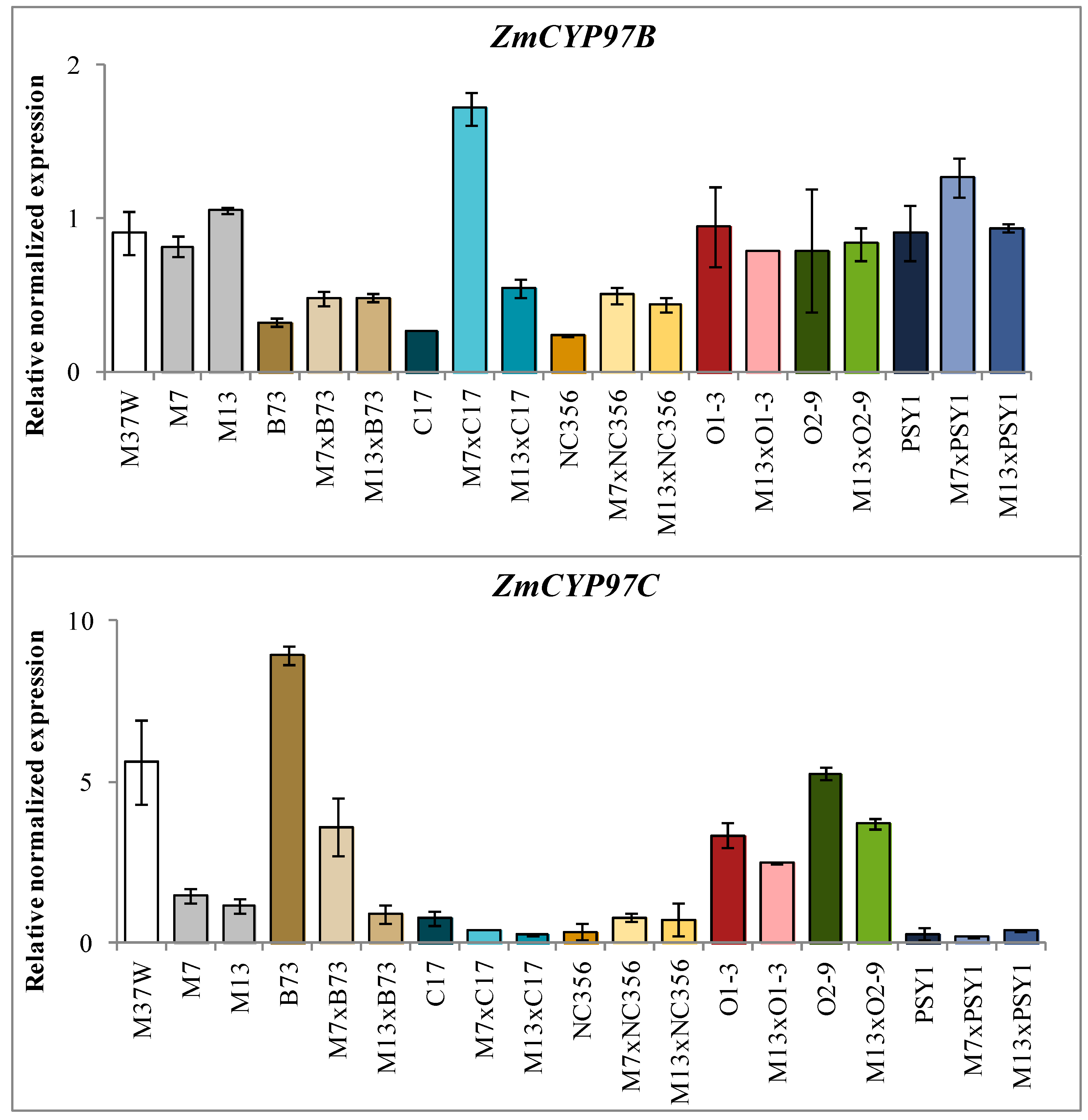
2.3. Analysis of Transgene and Endogenous Gene Expression in Hybrids Derived from BCH-Silenced Parents
3. Discussion
3.1. Silencing the Endogenous ZmBCH1 and ZmBCH2 Genes Causes β-Carotene to Accumulate in the Endosperm of Diverse Maize Hybrids
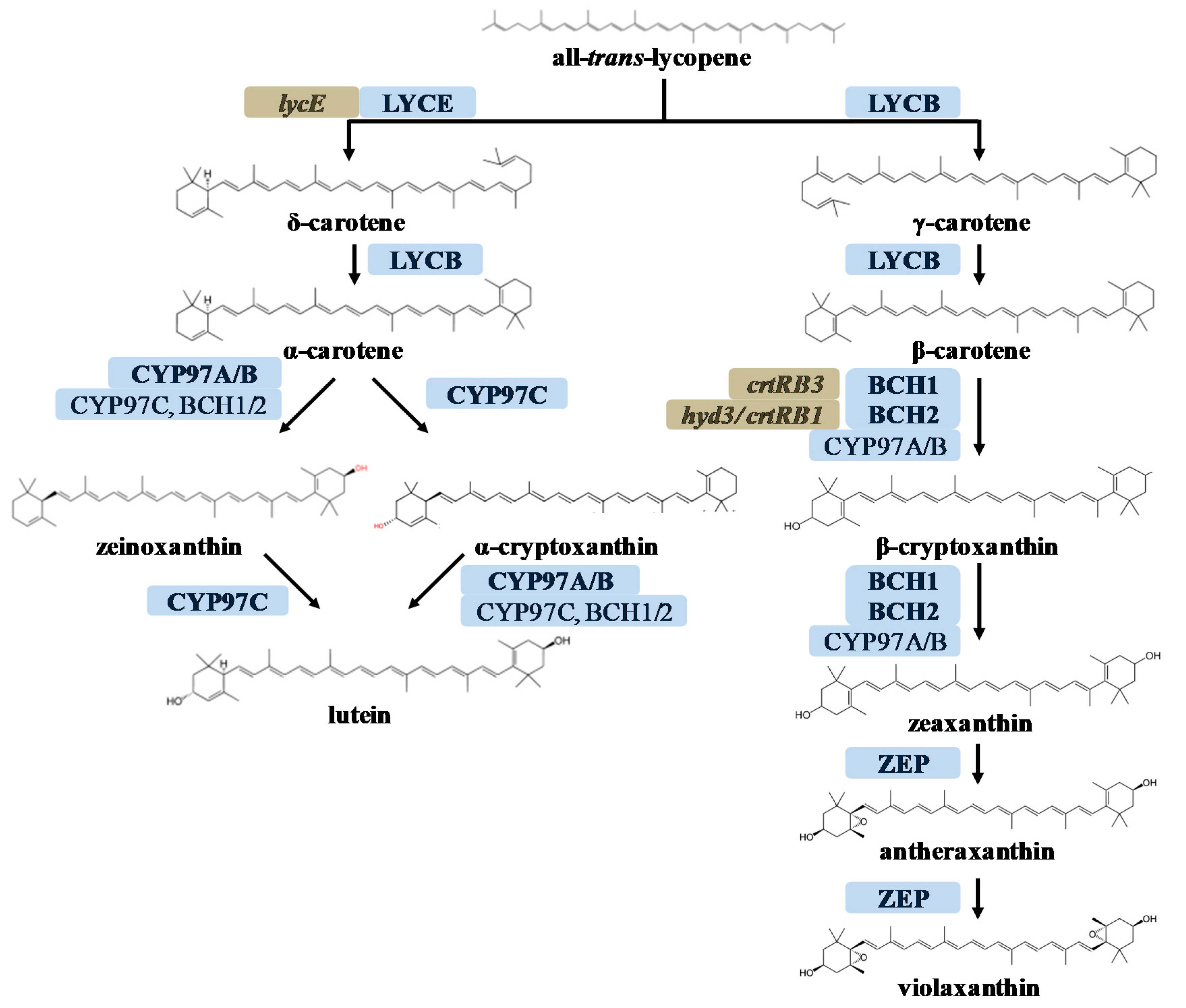
3.2. ZmBCH1 and ZmBCH2 Silencing Affects the Expression of CYP-Type ε-Hydroxylase Gene
4. Materials and Methods
4.1. Gene Cloning and Vector Construction
4.2. Maize Transformation and Plant Growth
4.3. RNA Extraction and Expression Analysis
4.4. Carotenoid Extraction and UPLC (Ultra Performance Liquid Chromatography) Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bai, C.; Twyman, R.M.; Farré, G.; Sanahuja, G.; Christou, P.; Capell, T.; Zhu, C. A golden era—Pro-vitamin A enhancement in diverse crops. In Vitro Cell. Dev. Biol. Plant 2011, 47, 205–221. [Google Scholar] [CrossRef]
- Farré, G.; Bai, C.; Twyman, R.M.; Capell, T.; Christou, P.; Zhu, C. Nutritious crops producing multiple carotenoids—A metabolic balancing act. Trends Plant Sci. 2011, 16, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zorrilla-López, U.; Farré, G.; Zhu, C.; Sandmann, G.; Twyman, R.M.; Capell, T.; Christou, P. Nutritionally important carotenoids as consumer products. Phytochem. Rev. 2015, 14, 727–743. [Google Scholar] [CrossRef]
- Zhu, C.; Naqvi, S.; Gomez-Galera, S.; Pelacho, A.M.; Capell, T.; Christou, P. Transgenic strategies for the nutritional enhancement of plants. Trends Plant Sci. 2007, 12, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zhu, C.; Pérez-Massot, E.; Arjó, G.; Zorrilla-López, U.; Masip, G.; Banakar, R.; Sanahuja, G.; Farré, G.; Miralpeix, B.; et al. Can the world afford to ignore biotechnology solutions that address food insecurity? Plant Mol. Biol. 2013, 83, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Farré, G.; Sanahuja, G.; Naqvi, S.; Bai, C.; Capell, T.; Zhu, C.; Christou, P. Travel advice on the road to carotenoids in plants. Plant Sci. 2010, 179, 28–48. [Google Scholar] [CrossRef]
- Farré, G.; Blancquaert, D.; Capell, T.; Van Der Straeten, D.; Christou, P.; Zhu, C. Engineering complex metabolic pathways in plants. Annu. Rev. Plant Biol. 2014, 65, 187–223. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Concepción, M. Supply of precursors for carotenoid biosynthesis in plants. Arch. Biochem. Biophys. 2010, 504, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Smith, J.J.; Tian, L.; DellaPenna, D. The evolution and function of carotenoid hydroxylases in Arabidopsis. Plant Cell Physiol. 2009, 50, 463–479. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Cheng, K.M.; Craft, N.E.; Hamberger, B.; Douglas, C.J. Over-expression of Arabidopsis thaliana carotenoid hydroxylases individually and in combination with a beta-carotene ketolase provides insight into in vivo functions. Phytochemistry 2010, 71, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Galpaz, N.; Ronen, G.; Khalfa, Z.; Zamir, D.; Hirschberg, J. A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 2006, 18, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosio, C.; Stigliani, A.L.; Giorio, G. Overexpression of CrtR-b2 (carotene β hydroxylase 2) from S. lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Res. 2011, 20, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Stigliani, A.L.; Giorio, G.; D’Ambrosio, C. Characterization of P450 carotenoid β- and epsilon-hydroxylases of tomato and transcriptional regulation of xanthophyll biosynthesis in root, leaf, petal and fruit. Plant Cell Physiol. 2011, 52, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, R.F.; Jaradat, T.T.; Wurtzel, E.T. Escherichia coli as a platform for functional expression of plant P450 carotene hydroxylases. Arch. Biochem. Biophys. 2007, 458, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.Z.; Chao, D.Y.; Shan, J.X.; Zhu, M.Z.; Shi, M.; Gao, J.P.; Lin, H.X. Rice carotenoid β-ring hydroxylase CYP97A4 is involved in lutein biosynthesis. Plant Cell Physiol. 2012, 53, 987–1002. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, R.; Gallagher, C.E.; Licciardello, N.; Cuttriss, A.J.; Quinlan, R.F.; Wurtzel, E.T. Metabolite sorting of a germplasm collection reveals the hydroxylase 3 locus as a new target for maize provitamin A biofortification. Plant Physiol. 2009, 151, 1635–1645. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Farre, G.; Naqvi, S.; Breitenbach, J.; Sanahuja, G.; Bai, C.; Sandmann, G.; Capell, T.; Christou, P.; Zhu, C. Cloning and functional characterization of the maize carotenoid isomerase and β-carotene hydroxylase genes and their regulation during endosperm maturation. Transgenic Res. 2010, 19, 1053–1068. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Kandianis, C.B.; Harjes, C.E.; Bai, L.; Kim, E.; Yang, X.; Skinner, D.J.; Fu, Z.; Mitchell, S.; Li, Q.; et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 2010, 42, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Han, Y.; Li, Z.; Fu, Y.; Fu, Z.; Xu, S.; Li, J.; Yan, J.; Yang, X. ZmcrtRB3 encodes a carotenoid hydroxylase that affects the accumulation of α-carotene in maize kernel. J. Integr. Plant Biol. 2012, 54, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Berman, J.; Sheng, Y.; Wang, Y.; Capell, T.; Shi, L.; Ni, X.; Sandmann, G.; Christou, P.; Zhu, C. Cloning and functional characterization of the maize (Zea mays L.) carotenoid epsilon hydroxylase gene. PLoS ONE 2015, 10, e0128758. [Google Scholar] [CrossRef] [PubMed]
- Babu, R.; Palacios Rojas, N.; Gao, S.; Yan, J.; Prixley, K. Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 2013, 126, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Owens, B.F.; Lipka, A.E.; Magallanes-Lundback, M.; Tiede, T.; Diepenbrock, C.H.; Kandianis, C.B.; Kim, E.; Cepela, J.; Mateos-Hernandez, M.; Buell, C.R.; et al. A foundation for provitamin A biofortification of maize: Genome-wide association and genomic prediction models of carotenoid levels. Genetics 2014, 198, 1699–1716. [Google Scholar] [CrossRef] [PubMed]
- Van Eck, J.; Conlin, B.; Garvin, D.F.; Mason, H.; Navarre, D.; Brown, C.R. Enhancing beta-carotene content in potato by RNAi-mediated silencing of the beta-carotene hydroxylase gene. Am. J. Potato Res. 2007, 84, 331–342. [Google Scholar] [CrossRef]
- Diretto, G.; Welsch, R.; Tavazza, R.; Mourgues, F.; Pizzichini, D.; Beyer, P.; Giuliano, G. Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol. 2007, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Pons, E.; Alquézar, B.; Rodríguez, A.; Martorell, P.; Genovés, S.; Ramón, D.; Rodrigo, M.J.; Zacarías, L.; Peña, L. Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 2014, 12, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Wang, X.T.; Miao, Y.J.; Wang, C.; Zang, M.L.; Chen, X.; Li, M.; Li, X.; Wang, Q.; Li, K.; et al. Metabolic engineering of wheat provitamin A by simultaneously overexpressing CrtB and silencing carotenoid hydroxylase (TaHYD). J. Agric. Food Chem. 2015, 63, 9083–9092. [Google Scholar] [CrossRef] [PubMed]
- Harjes, C.E.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T.; et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Naqvi, S.; Breitenbach, J.; Sandmann, G.; Christou, P.; Capell, T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 18232–18237. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, S.; Zhu, C.; Farre, G.; Sandmann, G.; Capell, T.; Christou, P. Synergistic metabolism in hybrid corn indicates bottlenecks in the carotenoid pathway and leads to the accumulation of extraordinary levels of the nutritionally important carotenoid zeaxanthin. Plant Biotechnol. J. 2011, 9, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zorrilla-López, U.; Medina, V.; Farré, G.; Sandmann, G.; Capell, T.; Christou, P.; Zhu, C. The Arabidopsis ORANGE (AtOR) gene promotes carotenoid accumulation in transgenic corn hybrids derived from parental lines with limited carotenoid pools. Plant Cell Rep. 2017, 36, 933–945. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berman, J.; Zorrilla-López, U.; Sandmann, G.; Capell, T.; Christou, P.; Zhu, C. The Silencing of Carotenoid β-Hydroxylases by RNA Interference in Different Maize Genetic Backgrounds Increases the β-Carotene Content of the Endosperm. Int. J. Mol. Sci. 2017, 18, 2515. https://doi.org/10.3390/ijms18122515
Berman J, Zorrilla-López U, Sandmann G, Capell T, Christou P, Zhu C. The Silencing of Carotenoid β-Hydroxylases by RNA Interference in Different Maize Genetic Backgrounds Increases the β-Carotene Content of the Endosperm. International Journal of Molecular Sciences. 2017; 18(12):2515. https://doi.org/10.3390/ijms18122515
Chicago/Turabian StyleBerman, Judit, Uxue Zorrilla-López, Gerhard Sandmann, Teresa Capell, Paul Christou, and Changfu Zhu. 2017. "The Silencing of Carotenoid β-Hydroxylases by RNA Interference in Different Maize Genetic Backgrounds Increases the β-Carotene Content of the Endosperm" International Journal of Molecular Sciences 18, no. 12: 2515. https://doi.org/10.3390/ijms18122515