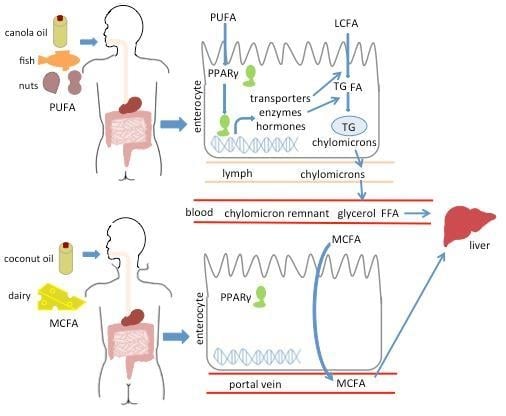
PPARγ Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium
Abstract
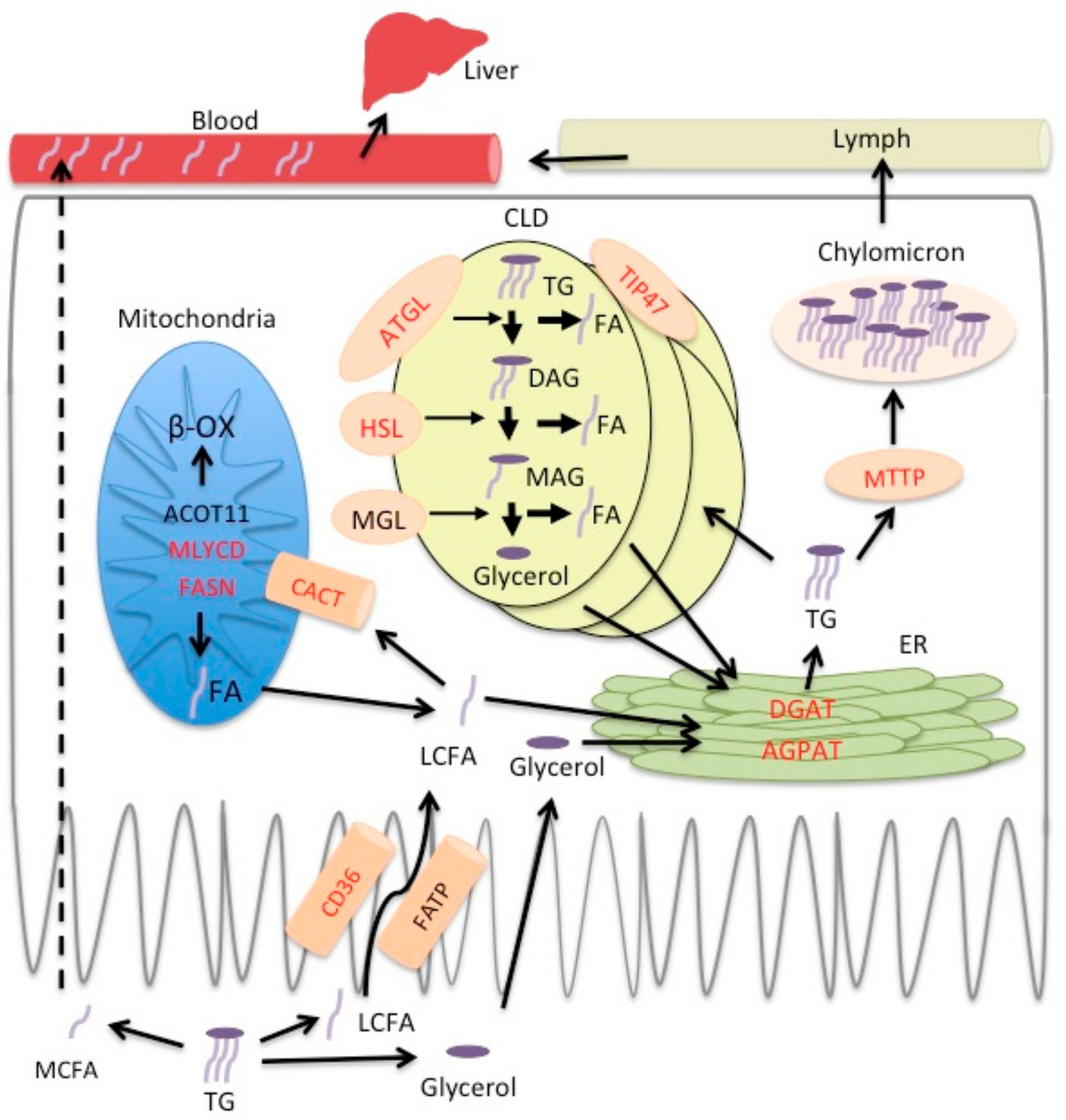
:1. Introduction
2. Results
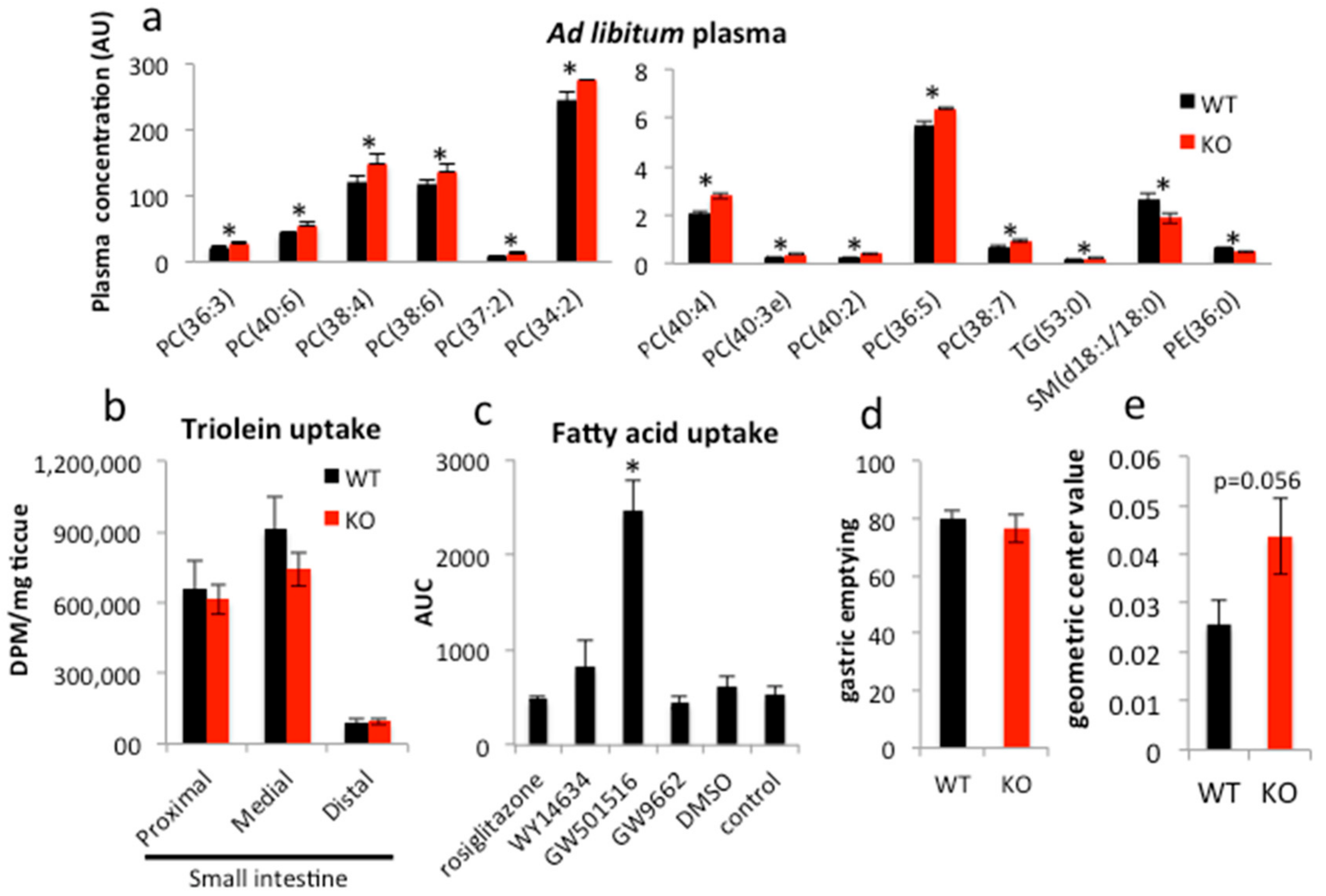
2.1. PPARγ Regulates Lipid Transit but Not Uptake in Small Intestine
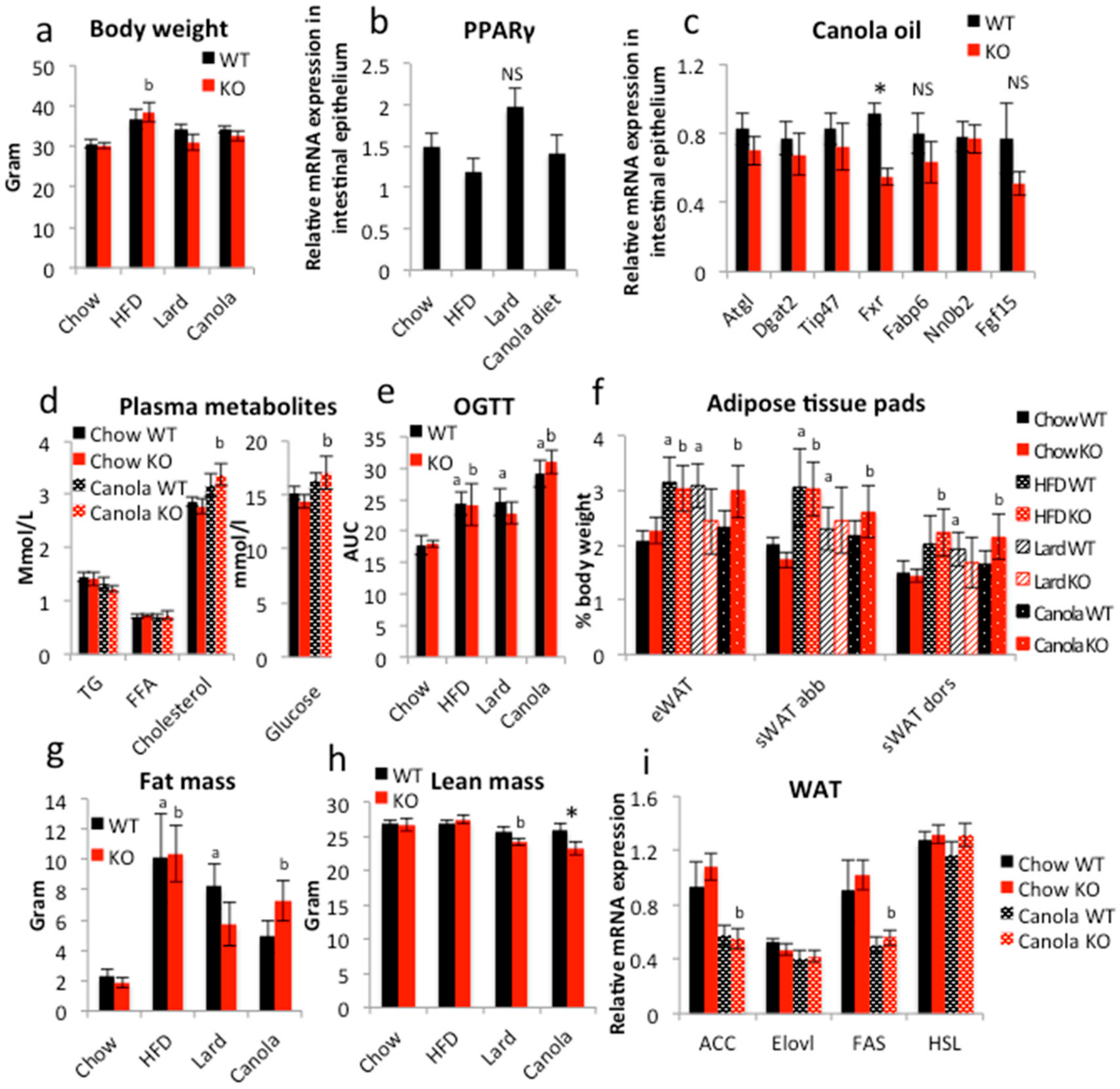
2.2. Long-Term Canola Oil Diet Results in Modest Body Composition Changes in iePPARγKO Compared to WT Mice
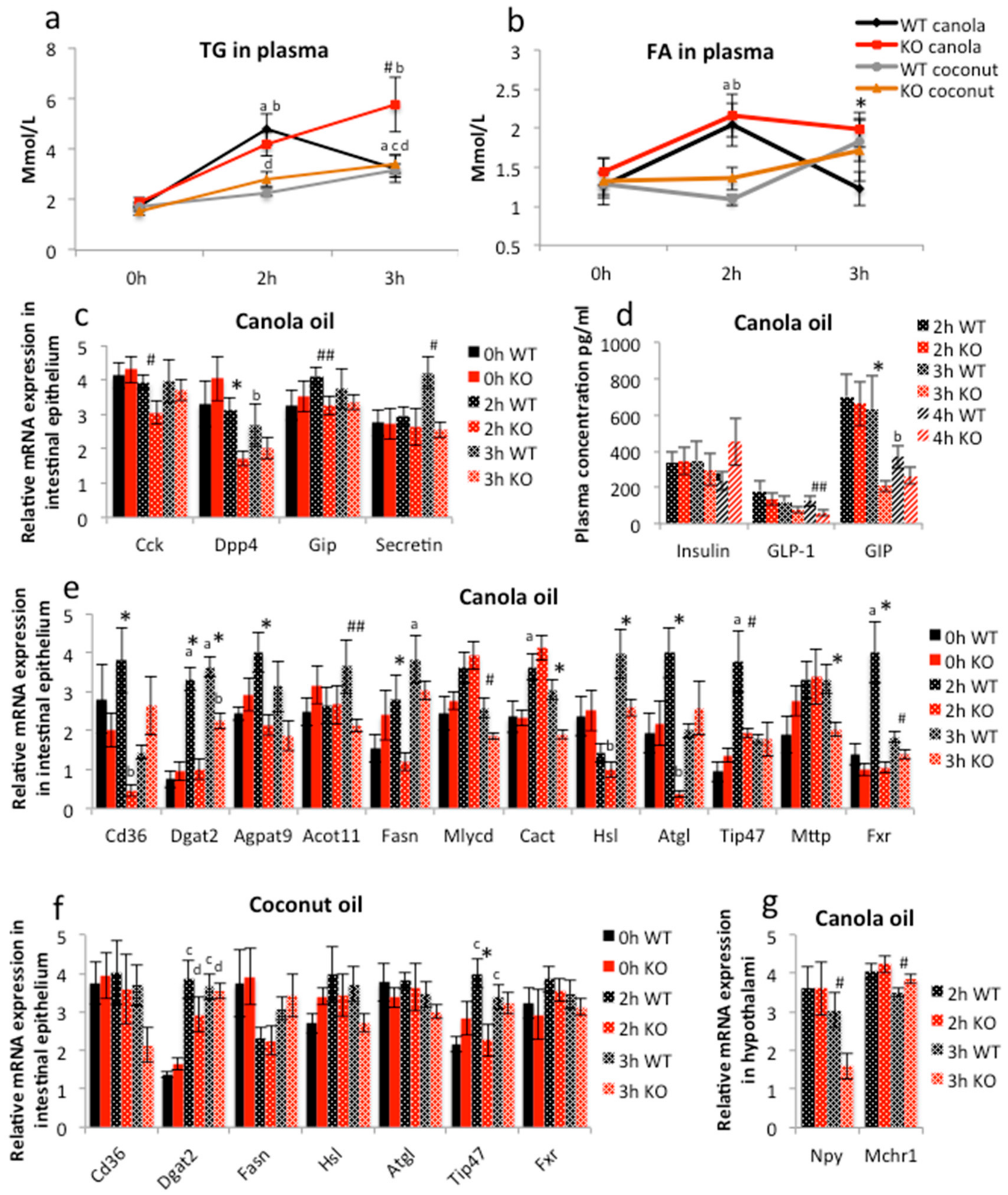
2.3. PPARγ Affects Lipid Metabolism in Duodenal Enterocytes
3. Discussion
4. Materials and Methods
4.1. Mouse Handling
4.2. Intestinal 3H-Triolein
4.3. Gastric Emptying and Intestinal Motility
4.4. RT-qPCR
4.5. Plasma Analysis
4.6. Cell Culture
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hofmann, A.F.; Borgstrom, B. Hydrolysis of long-chain monoglycerides in micellar solution by pancreatic lipase. Biochim. Biophys. Acta 1963, 70, 317–731. [Google Scholar] [CrossRef]
- Iqbal, J.; Hussain, M.M. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1183–E1194. [Google Scholar] [CrossRef] [PubMed]
- Chabowski, A.; Gorski, J.; Luiken, J.J.; Glatz, J.F.; Bonen, A. Evidence for concerted action of FAT/CD36 and FABPpm to increase fatty acid transport across the plasma membrane. Prostaglandins Leukot. Essent. Fatty Acids 2007, 77, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.E.; Lodish, H.F. Expression cloning and characterization of a novel adipocyte long chain fatty acid transport protein. Cell 1994, 79, 427–436. [Google Scholar] [CrossRef]
- Coleman, R.A.; Haynes, E.B. Monoacylglycerol acyltransferase. Evidence that the activities from rat intestine and suckling liver are tissue-specific isoenzymes. J. Biol. Chem. 1986, 261, 224–228. [Google Scholar] [PubMed]
- Yen, C.L.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V., Jr. Thematic review series: Glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [PubMed]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef] [PubMed]
- Black, D.D. Development and physiological regulation of intestinal lipid absorption. I. Development of intestinal lipid absorption: Cellular events in chylomicron assembly and secretion. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G519–G524. [Google Scholar] [PubMed]
- Mansbach, C.M., 2nd; Gorelick, F. Development and physiological regulation of intestinal lipid absorption. II. Dietary lipid absorption, complex lipid synthesis, and the intracellular packaging and secretion of chylomicrons. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 293, G645–G650. [Google Scholar]
- Zhu, J.; Lee, B.; Buhman, K.K.; Cheng, J.X. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J. Lipid Res. 2009, 50, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Grober, J.; Lucas, S.; Sorhede-Winzell, M.; Zaghini, I.; Mairal, A.; Contreras, J.A.; Besnard, P.; Holm, C.; Langin, D. Hormone-sensitive lipase is a cholesterol esterase of the intestinal mucosa. J. Biol. Chem. 2003, 278, 6510–6515. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, G.; Lass, A.; Zimmermann, R.; Gorkiewicz, G.; Meyer, C.; Rozman, J.; Heldmaier, G.; Maier, R.; Theussl, C.; Eder, S.; et al. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 2006, 312, 734–737. [Google Scholar] [CrossRef] [PubMed]
- Eyres, L.; Eyres, M.F.; Chisholm, A.; Brown, R.C. Coconut oil consumption and cardiovascular risk factors in humans. Nutr. Rev. 2016, 74, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Bach, A.C.; Babayan, V.K. Medium-chain triglycerides: An update. Am. J. Clin. Nutr. 1982, 36, 950–962. [Google Scholar] [PubMed]
- Orsavova, J.; Misurcova, L.; Ambrozova, J.V.; Vicha, R.; Mlcek, J. Fatty Acids Composition of Vegetable Oils and Its Contribution to Dietary Energy Intake and Dependence of Cardiovascular Mortality on Dietary Intake of Fatty Acids. Int. J. Mol. Sci. 2015, 16, 12871–12890. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, H.; Ollilainen, V.; Piironen, V.; Lampi, A.M. Tocopherol, tocotrienol and plant sterol contents of vegetable oils and industrial fats. J. Food Compos. Anal. 2008, 21, 152–161. [Google Scholar] [CrossRef]
- Lin, L.; Allemekinders, H.; Dansby, A.; Campbell, L.; Durance-Tod, S.; Berger, A.; Jones, P.J. Evidence of health benefits of canola oil. Nutr. Rev. 2013, 71, 370–385. [Google Scholar] [CrossRef] [PubMed]
- Michalik, L.; Auwerx, J.; Berger, J.P.; Chatterjee, V.K.; Glass, C.K.; Gonzalez, F.J.; Grimaldi, P.A.; Kadowaki, T.; Lazar, M.A.; O’Rahilly, S.; et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol. Rev. 2006, 58, 726–741. [Google Scholar] [PubMed]
- De Vogel-van den Bosch, H.M.; Bunger, M.; de Groot, P.J.; Bosch-Vermeulen, H.; Hooiveld, G.J.; Muller, M. PPARα-mediated effects of dietary lipids on intestinal barrier gene expression. BMC Genom. 2008, 9. [Google Scholar] [CrossRef] [PubMed]
- Bunger, M.; van den Bosch, H.M.; van der Meijde, J.; Kersten, S.; Hooiveld, G.J.; Muller, M. Genome-wide analysis of PPARα activation in murine small intestine. Physiol. Genom. 2007, 30, 192–204. [Google Scholar] [CrossRef] [PubMed]
- Varnat, F.; Heggeler, B.B.; Grisel, P.; Boucard, N.; Corthesy-Theulaz, I.; Wahli, W.; Desvergne, B. PPARβ/delta regulates paneth cell differentiation via controlling the hedgehog signaling pathway. Gastroenterology 2006, 131, 538–553. [Google Scholar] [CrossRef] [PubMed]
- Poirier, H.; Niot, I.; Monnot, M.C.; Braissant, O.; Meunier-Durmort, C.; Costet, P.; Pineau, T.; Wahli, W.; Willson, T.M.; Besnard, P. Differential involvement of peroxisome-proliferator-activated receptors α and delta in fibrate and fatty-acid-mediated inductions of the gene encoding liver fatty-acid-binding protein in the liver and the small intestine. Biochem. J. 2001, 355, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Byndloss, M.X.; Olsan, E.E.; Rivera-Chavez, F.; Tiffany, C.R.; Cevallos, S.A.; Lokken, K.L.; Torres, T.P.; Byndloss, A.J.; Faber, F.; Gao, Y.; et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 2017, 357, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Escher, P.; Braissant, O.; Basu-Modak, S.; Michalik, L.; Wahli, W.; Desvergne, B. Rat PPARs: Quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology 2001, 142, 4195–4202. [Google Scholar] [CrossRef] [PubMed]
- Harmon, G.S.; Dumlao, D.S.; Ng, D.T.; Barrett, K.E.; Dennis, E.A.; Dong, H.; Glass, C.K. Pharmacological correction of a defect in PPAR-γ signaling ameliorates disease severity in Cftr-deficient mice. Nat. Med. 2010, 16, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Mansen, A.; Guardiola-Diaz, H.; Rafter, J.; Branting, C.; Gustafsson, J.A. Expression of the peroxisome proliferator-activated receptor (PPAR) in the mouse colonic mucosa. Biochem. Biophys. Res. Commun. 1996, 222, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Braissant, O.; Foufelle, F.; Scotto, C.; Dauca, M.; Wahli, W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology 1996, 137, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Marion-Letellier, R.; Dechelotte, P.; Iacucci, M.; Ghosh, S. Dietary modulation of peroxisome proliferator-activated receptor γ. Gut 2009, 58, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Willson, T.M.; Wahli, W. Peroxisome proliferator-activated receptor agonists. Curr. Opin. Chem. Biol. 1997, 1, 235–241. [Google Scholar] [CrossRef]
- Bassaganya-Riera, J.; Hontecillas, R. CLA and n-3 PUFA differentially modulate clinical activity and colonic PPAR-responsive gene expression in a pig model of experimental IBD. Clin. Nutr. 2006, 25, 454–465. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Lichtenstein, G.R.; Deren, J.J.; Sands, B.E.; Hanauer, S.B.; Katz, J.A.; Lashner, B.; Present, D.H.; Chuai, S.; Ellenberg, J.H.; et al. Rosiglitazone for active ulcerative colitis: A randomized placebo-controlled trial. Gastroenterology 2008, 134, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Lichtenstein, G.R.; Stein, R.B.; Deren, J.J.; Judge, T.A.; Fogt, F.; Furth, E.E.; Demissie, E.J.; Hurd, L.B.; Su, C.G.; et al. An open-label trial of the PPAR-γ ligand rosiglitazone for active ulcerative colitis. Am. J. Gastroenterol. 2001, 96, 3323–3328. [Google Scholar] [PubMed]
- Sanchez-Hidalgo, M.; Martin, A.R.; Villegas, I.; de la Lastra, C.A. Rosiglitazone, a PPARγ ligand, modulates signal transduction pathways during the development of acute TNBS-induced colitis in rats. Eur. J. Pharmacol. 2007, 562, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Shah, Y.M.; Morimura, K.; Gonzalez, F.J. Expression of peroxisome proliferator-activated receptor-γ in macrophage suppresses experimentally induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G657–G666. [Google Scholar] [CrossRef] [PubMed]
- Su, C.G.; Wen, X.; Bailey, S.T.; Jiang, W.; Rangwala, S.M.; Keilbaugh, S.A.; Flanigan, A.; Murthy, S.; Lazar, M.A.; Wu, G.D. A novel therapy for colitis utilizing PPAR-γ ligands to inhibit the epithelial inflammatory response. J. Clin Investig. 1999, 104, 383–389. [Google Scholar] [CrossRef] [PubMed]
- Rousseaux, C.; Lefebvre, B.; Dubuquoy, L.; Lefebvre, P.; Romano, O.; Auwerx, J.; Metzger, D.; Wahli, W.; Desvergne, B.; Naccari, G.C.; et al. Intestinal antiinflammatory effect of 5-aminosalicylic acid is dependent on peroxisome proliferator-activated receptor-γ. J. Exp. Med. 2005, 201, 1205–1215. [Google Scholar] [CrossRef] [PubMed]
- Wahli, W. A gut feeling of the PXR, PPAR and NF-κB connection. J. Intern. Med. 2008, 263, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Cerbone, A.; Toaldo, C.; Laurora, S.; Briatore, F.; Pizzimenti, S.; Dianzani, M.U.; Ferretti, C.; Barrera, G. 4-Hydroxynonenal and PPARγ ligands affect proliferation, differentiation, and apoptosis in colon cancer cells. Free Radic. Biol. Med. 2007, 42, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Martinasso, G.; Oraldi, M.; Trombetta, A.; Maggiora, M.; Bertetto, O.; Canuto, R.A.; Muzio, G. Involvement of PPARs in Cell Proliferation and Apoptosis in Human Colon Cancer Specimens and in Normal and Cancer Cell Lines. PPAR Res. 2007, 2007. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.; Pradeep, A.; Wong, L.; Rana, A.; Rana, B. Peroxisome proliferator-activated receptor γ activation can regulate β-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 2004, 279, 35583–35594. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.P.; Zhang, X.; Xie, W.F. Differentiation therapy for solid tumors. J. Dig. Dis. 2014, 15, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Sheng, H.; DuBois, R.N. Peroxisome proliferator-activated receptors modulate K-Ras-mediated transformation of intestinal epithelial cells. Cancer Res. 2002, 62, 3282–3288. [Google Scholar] [PubMed]
- Sato, N.; Kozar, R.A.; Zou, L.; Weatherall, J.M.; Attuwaybi, B.; Moore-Olufemi, S.D.; Weisbrodt, N.W.; Moore, F.A. Peroxisome proliferator-activated receptor γ mediates protection against cyclooxygenase-2-induced gut dysfunction in a rodent model of mesenteric ischemia/reperfusion. Shock 2005, 24, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Duszka, K.; Picard, A.; Ellero-Simatos, S.; Chen, J.; Defernez, M.; Paramalingam, E.; Pigram, A.; Vanoaica, L.; Canlet, C.; Parini, P.; et al. Intestinal PPARγ signalling is required for sympathetic nervous system activation in response to caloric restriction. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Anghel, S.I.; Wahli, W. Fat poetry: A kingdom for PPAR γ. Cell Res. 2007, 17, 486–511. [Google Scholar] [CrossRef] [PubMed]
- Brasaemle, D.L. Thematic review series: Adipocyte biology. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 2007, 48, 2547–2559. [Google Scholar] [PubMed]
- Macneil, D.J. The role of melanin-concentrating hormone and its receptors in energy homeostasis. Front. Endocrinol. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Shearman, L.P.; Camacho, R.E.; Sloan Stribling, D.; Zhou, D.; Bednarek, M.A.; Hreniuk, D.L.; Feighner, S.D.; Tan, C.P.; Howard, A.D.; van der Ploeg, L.H.; et al. Chronic MCH-1 receptor modulation alters appetite, body weight and adiposity in rats. Eur. J. Pharmacol. 2003, 475, 37–47. [Google Scholar] [CrossRef]
- Krey, G.; Braissant, O.; L’Horset, F.; Kalkhoven, E.; Perroud, M.; Parker, M.G.; Wahli, W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol. Endocrinol. 1997, 11, 779–791. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Viswakarma, N.; Batra, S.K.; Sambasiva Rao, M.; Reddy, J.K. Identification of promethin and PGLP as two novel up-regulated genes in PPARγ1-induced adipogenic mouse liver. Biochimie 2004, 86, 743–761. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.S.; Siersbaek, R.; Boergesen, M.; Nielsen, R.; Mandrup, S. Peroxisome proliferator-activated receptor γ and C/EBPα synergistically activate key metabolic adipocyte genes by assisted loading. Mol. Cell. Biol. 2014, 34, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Matsusue, K.; Kashireddy, P.; Cao, W.Q.; Yeldandi, V.; Yeldandi, A.V.; Rao, M.S.; Gonzalez, F.J.; Reddy, J.K. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J. Biol. Chem. 2003, 278, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.; Balard, P.; Coste, A.; Olagnier, D.; Lagane, C.; Authier, H.; Benoit-Vical, F.; Lepert, J.C.; Seguela, J.P.; Magnaval, J.F.; et al. IL-13 induces expression of CD36 in human monocytes through PPARγ activation. Eur. J. Immunol. 2007, 37, 1642–1652. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Shan, S.; Li, P.P.; Shen, Z.F.; Lu, X.P.; Cheng, J.; Ning, Z.Q. Peroxisome proliferator-activated receptor-γ transcriptionally up-regulates hormone-sensitive lipase via the involvement of specificity protein-1. Endocrinology 2006, 147, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, R.; Pedersen, T.A.; Hagenbeek, D.; Moulos, P.; Siersbaek, R.; Megens, E.; Denissov, S.; Borgesen, M.; Francoijs, K.J.; Mandrup, S.; et al. Genome-wide profiling of PPARγ: RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008, 22, 2953–2967. [Google Scholar] [CrossRef] [PubMed]
- Kershaw, E.E.; Schupp, M.; Guan, H.P.; Gardner, N.P.; Lazar, M.A.; Flier, J.S. PPARγ regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E1736–E1745. [Google Scholar] [CrossRef] [PubMed]
- Lapsys, N.M.; Kriketos, A.D.; Lim-Fraser, M.; Poynten, A.M.; Lowy, A.; Furler, S.M.; Chisholm, D.J.; Cooney, G.J. Expression of genes involved in lipid metabolism correlate with peroxisome proliferator-activated receptor γ expression in human skeletal muscle. J. Clin. Endocrinol. Metab. 2000, 85, 4293–4297. [Google Scholar] [CrossRef] [PubMed]
- Matsusue, K.; Haluzik, M.; Lambert, G.; Yim, S.H.; Gavrilova, O.; Ward, J.M.; Brewer, B., Jr.; Reitman, M.L.; Gonzalez, F.J. Liver-specific disruption of PPARγ in leptin-deficient mice improves fatty liver but aggravates diabetic phenotypes. J. Clin. Investig. 2003, 111, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Graugnard, D.E.; Piantoni, P.; Bionaz, M.; Berger, L.L.; Faulkner, D.B.; Loor, J.J. Adipogenic and energy metabolism gene networks in longissimus lumborum during rapid post-weaning growth in Angus and Angus x Simmental cattle fed high-starch or low-starch diets. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Young, M.E.; Goodwin, G.W.; Ying, J.; Guthrie, P.; Wilson, C.R.; Laws, F.A.; Taegtmeyer, H. Regulation of cardiac and skeletal muscle malonyl-CoA decarboxylase by fatty acids. Am. J. Physiol. Endocrinol. Metab. 2001, 280, E471–E479. [Google Scholar] [PubMed]
- Cao, J.; Li, J.L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA: Glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef] [PubMed]
- Meyer, B.M.; Werth, B.A.; Beglinger, C.; Hildebrand, P.; Jansen, J.B.; Zach, D.; Rovati, L.C.; Stalder, G.A. Role of cholecystokinin in regulation of gastrointestinal motor functions. Lancet 1989, 2, 12–15. [Google Scholar] [CrossRef]
- Harvey, R.F. Hormonal control of gastrointestinal motility. Am. J. Dig. Dis. 1975, 20, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Hammer, J.; Hammer, K.; Kletter, K. Lipids infused into the jejunum accelerate small intestinal transit but delay ileocolonic transit of solids and liquids. Gut 1998, 43, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Petit, V.; Arnould, L.; Martin, P.; Monnot, M.C.; Pineau, T.; Besnard, P.; Niot, I. Chronic high-fat diet affects intestinal fat absorption and postprandial triglyceride levels in the mouse. J. Lipid Res. 2007, 48, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.S.; Galligan, J.J.; Burks, T.F. Accurate measurement of intestinal transit in the rat. J. Pharmacol. Methods 1981, 6, 211–217. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duszka, K.; Oresic, M.; Le May, C.; König, J.; Wahli, W. PPARγ Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium. Int. J. Mol. Sci. 2017, 18, 2559. https://doi.org/10.3390/ijms18122559
Duszka K, Oresic M, Le May C, König J, Wahli W. PPARγ Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium. International Journal of Molecular Sciences. 2017; 18(12):2559. https://doi.org/10.3390/ijms18122559
Chicago/Turabian StyleDuszka, Kalina, Matej Oresic, Cedric Le May, Jürgen König, and Walter Wahli. 2017. "PPARγ Modulates Long Chain Fatty Acid Processing in the Intestinal Epithelium" International Journal of Molecular Sciences 18, no. 12: 2559. https://doi.org/10.3390/ijms18122559