Identification and Characterization of Hyphantria cunea Aminopeptidase N as a Binding Protein of Bacillus thuringiensis Cry1Ab35 Toxin
Abstract
:1. Introduction
2. Results
2.1. Toxicity of Cry1Ab35 Toxin against H. cunea
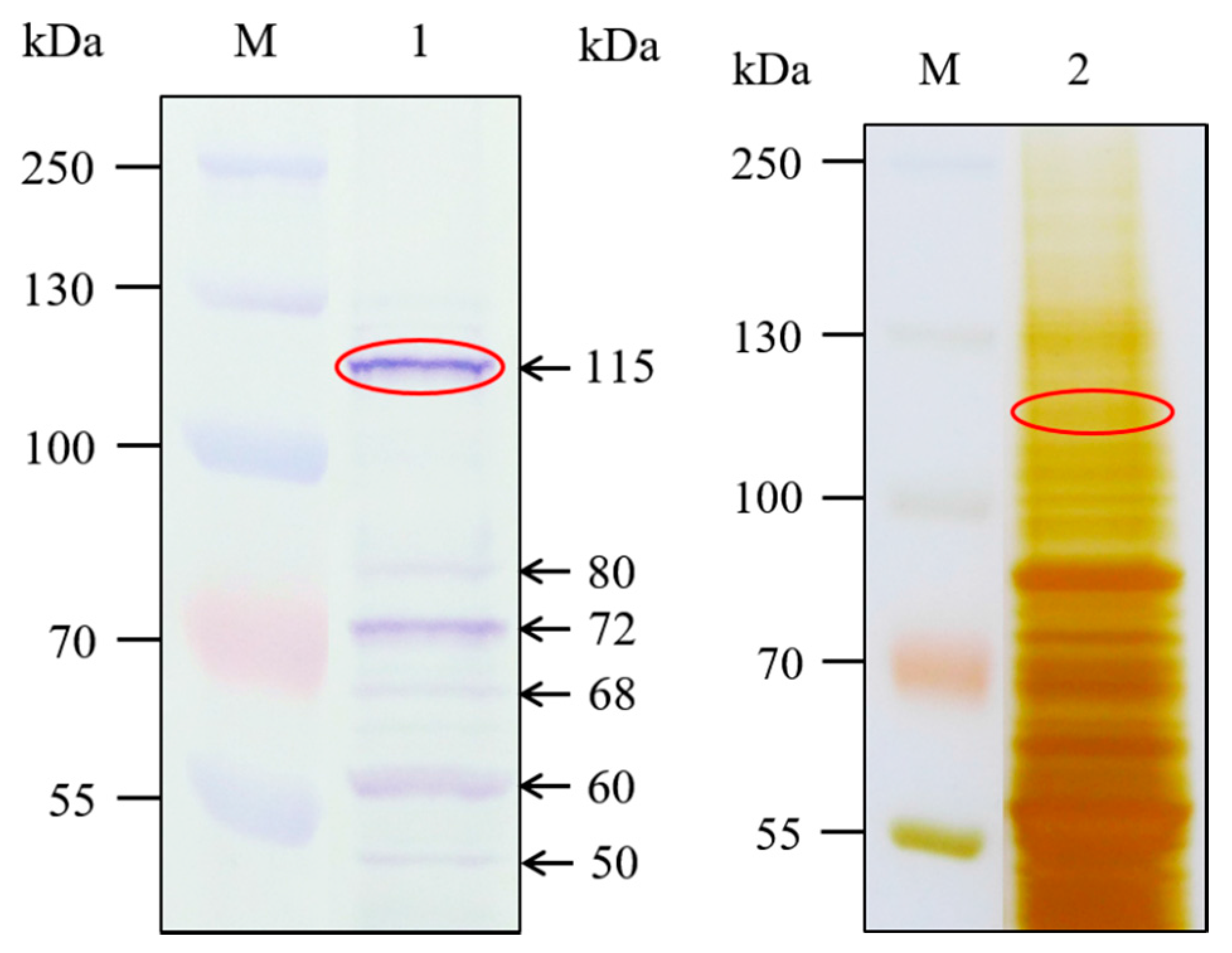
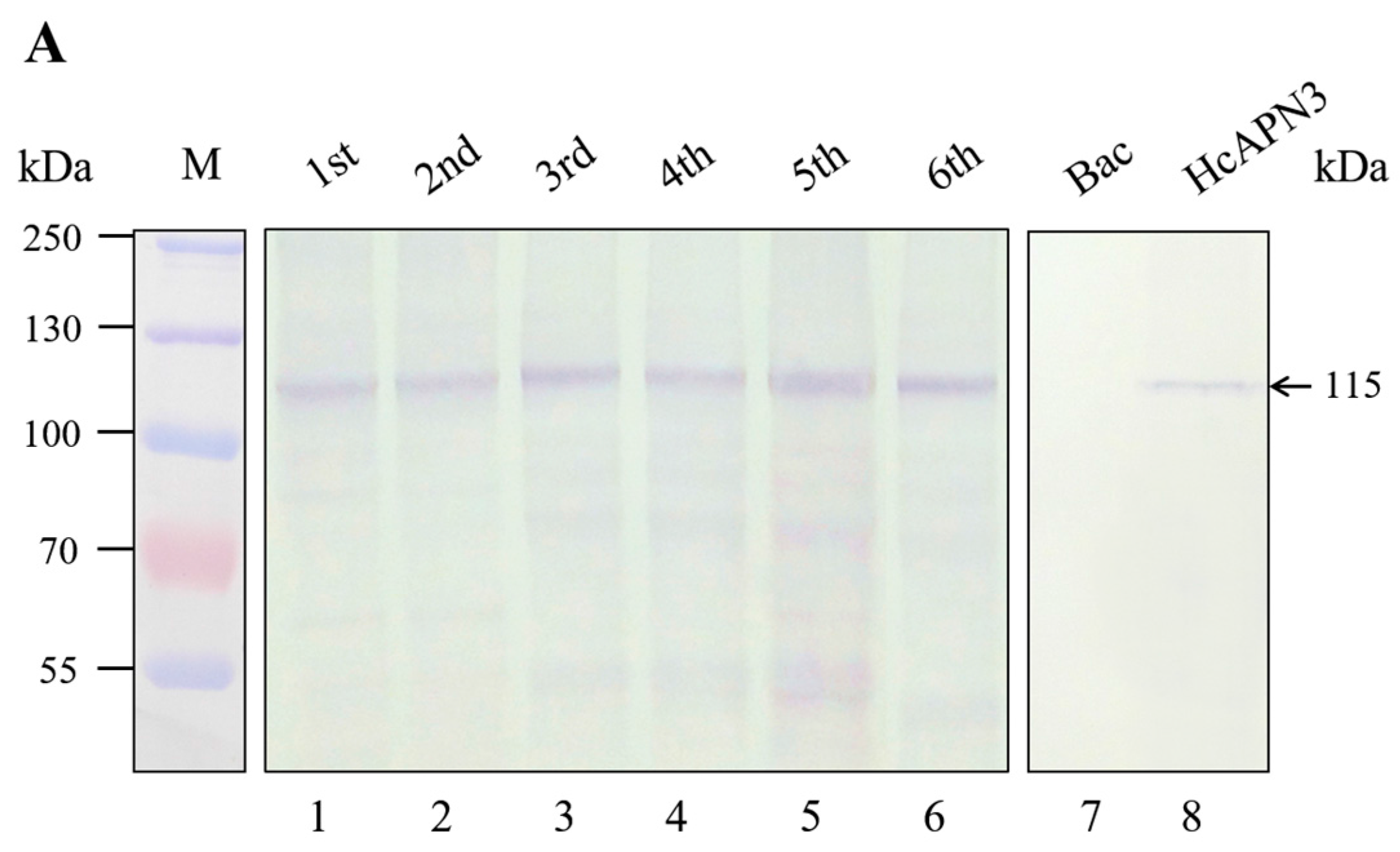
2.2. Detection of Cry1Ab-Binding Proteins in H. cunea BBMV
2.3. Cloning and Characterization of H. cunea HcAPN3
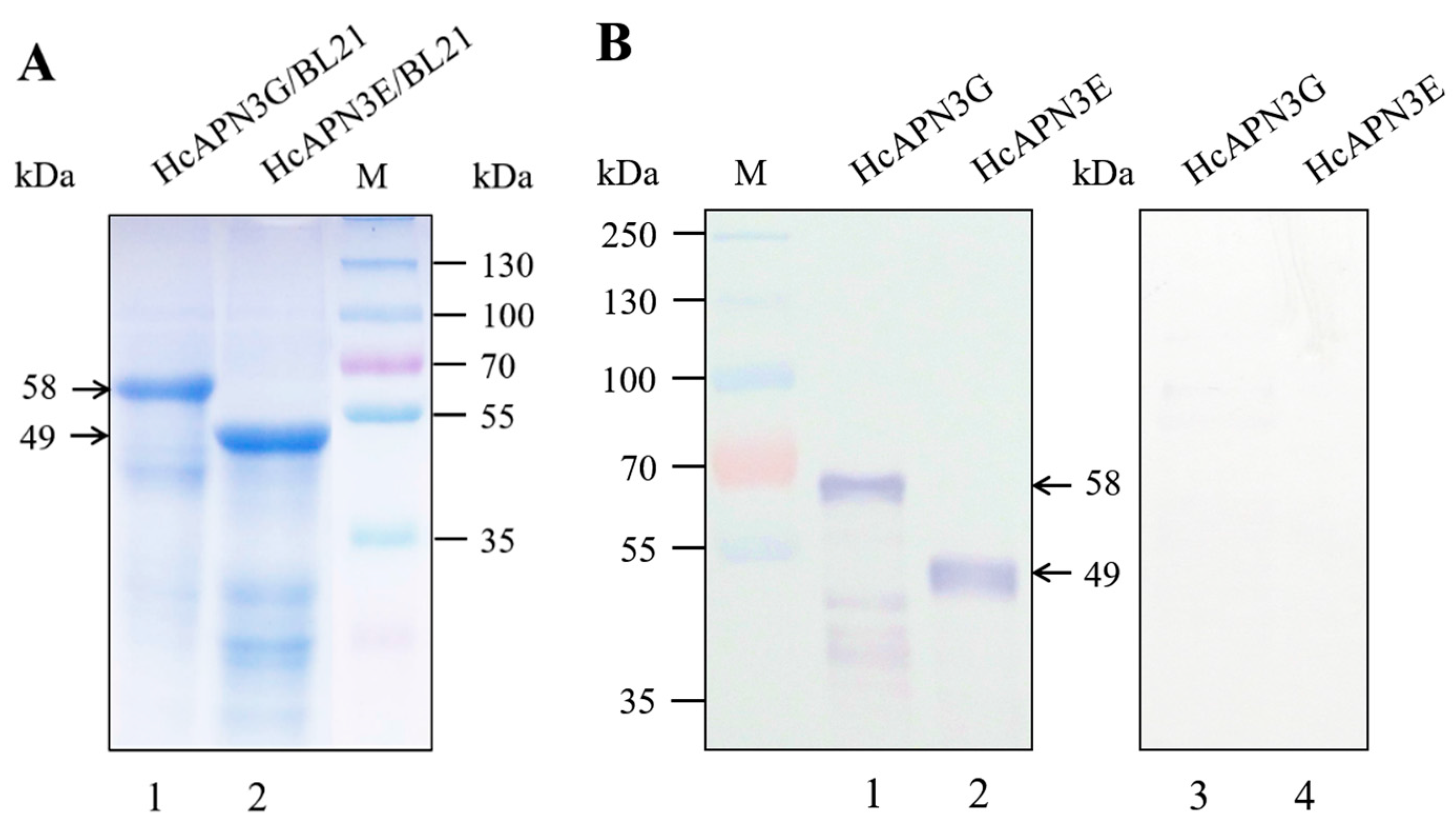
2.4. Expression of HcAPN3G and HcAPN3E and Their Binding to Cry1Ab35 Toxin
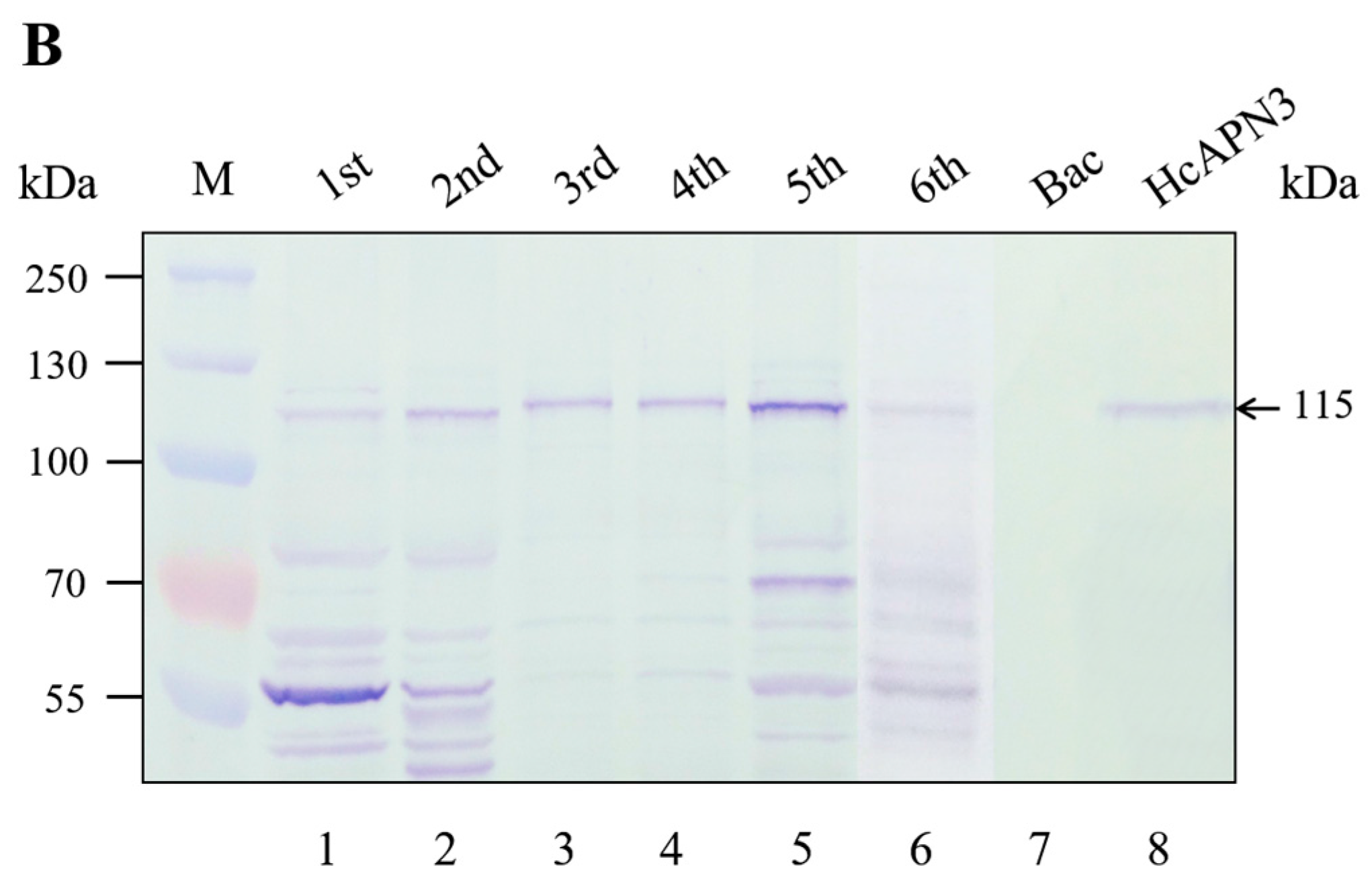
2.5. Binding of Cry1Ab35 Toxin to HcAPN3 Expressed in H. cunea BBMV and Infected Sf9 Cells
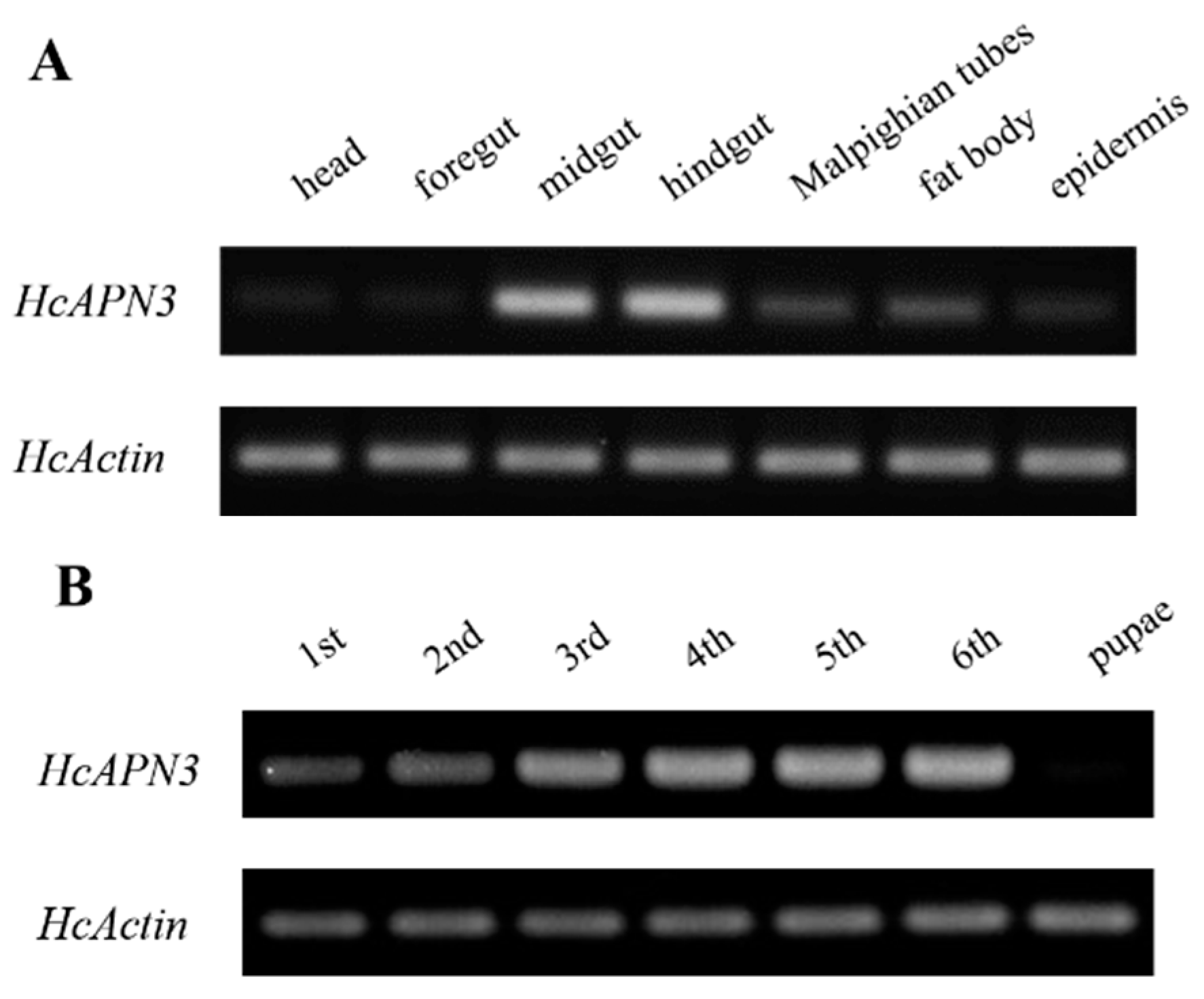
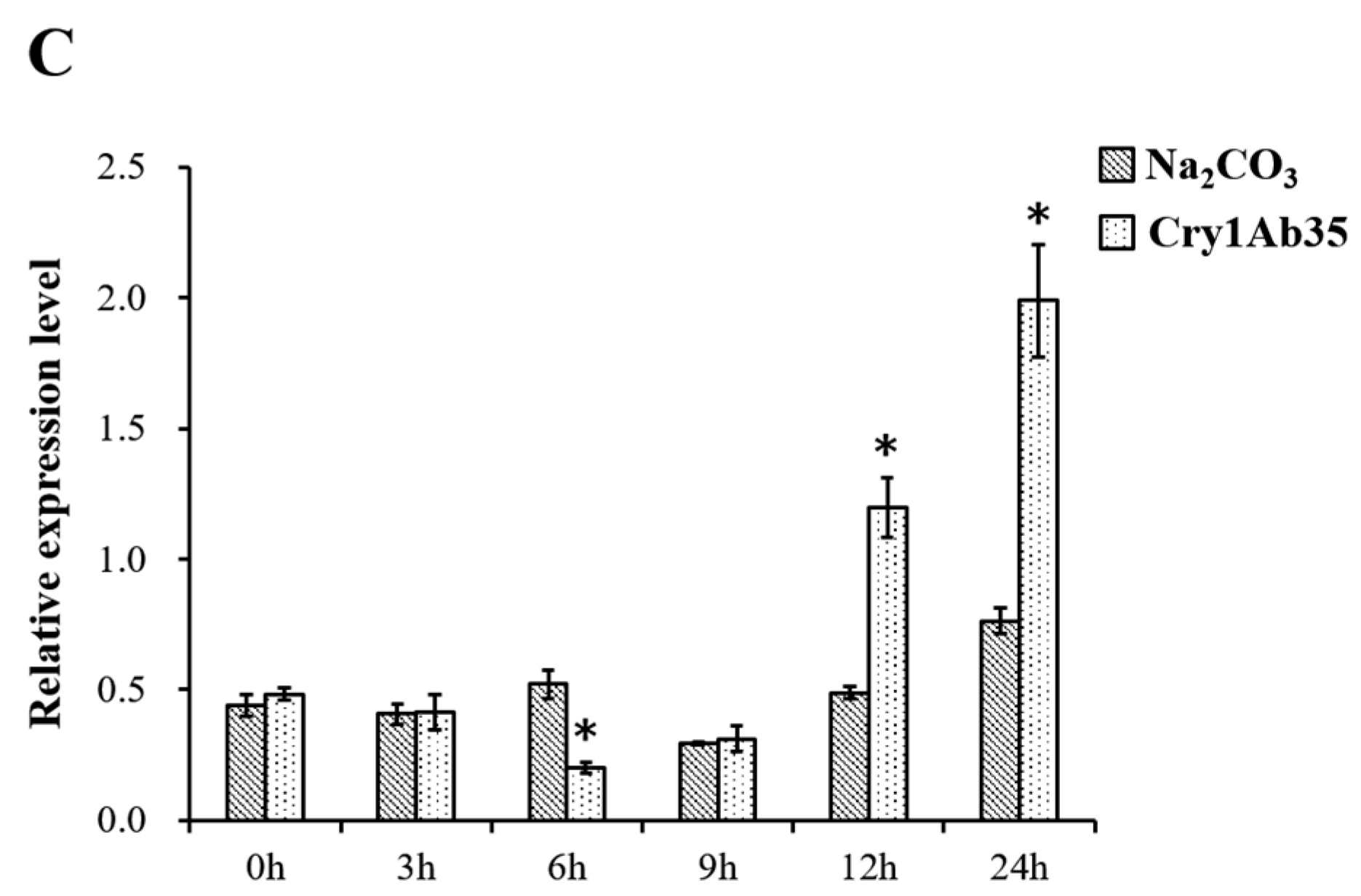
2.6. Spatiotemporal and Induced Expression Profiles of HcAPN3
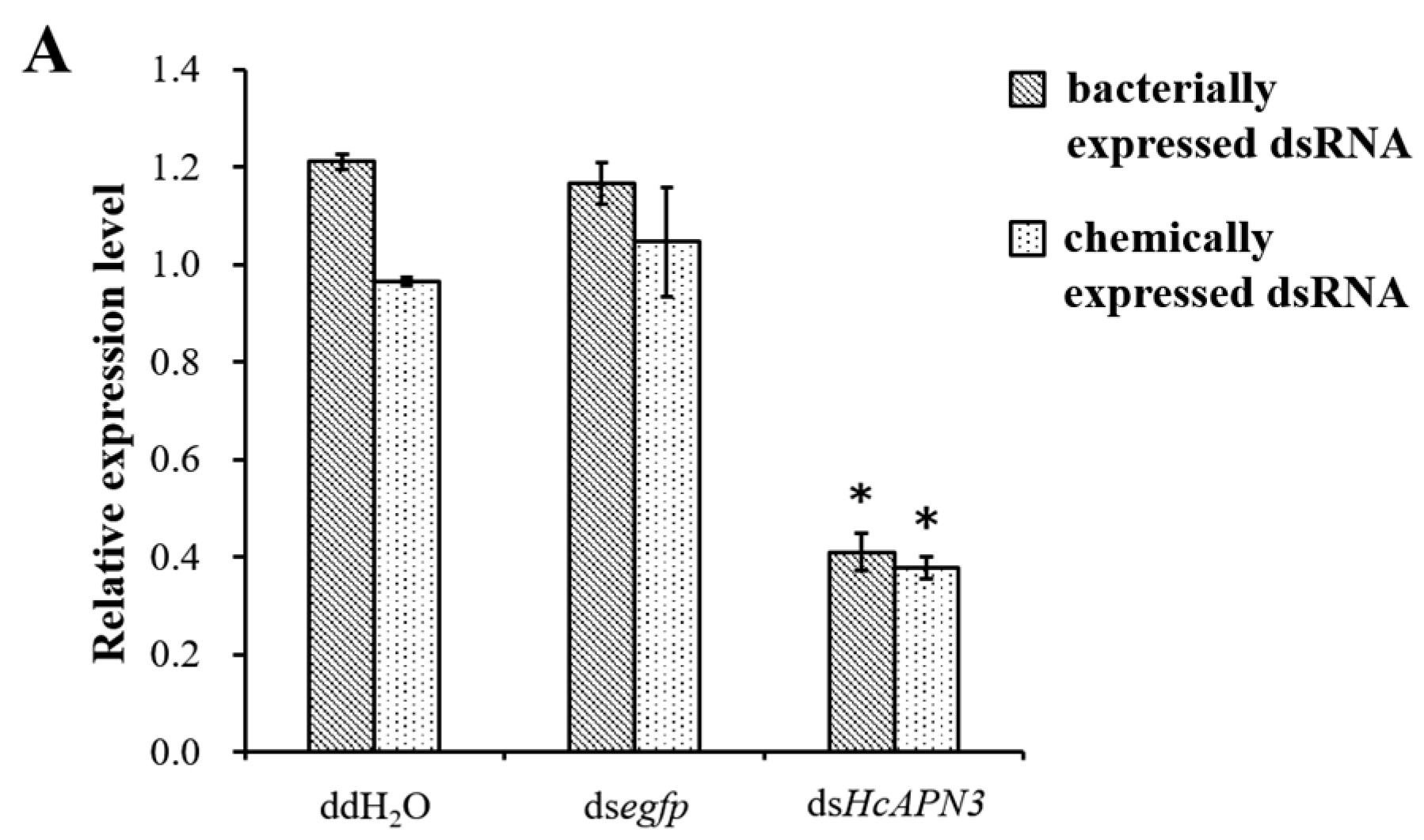
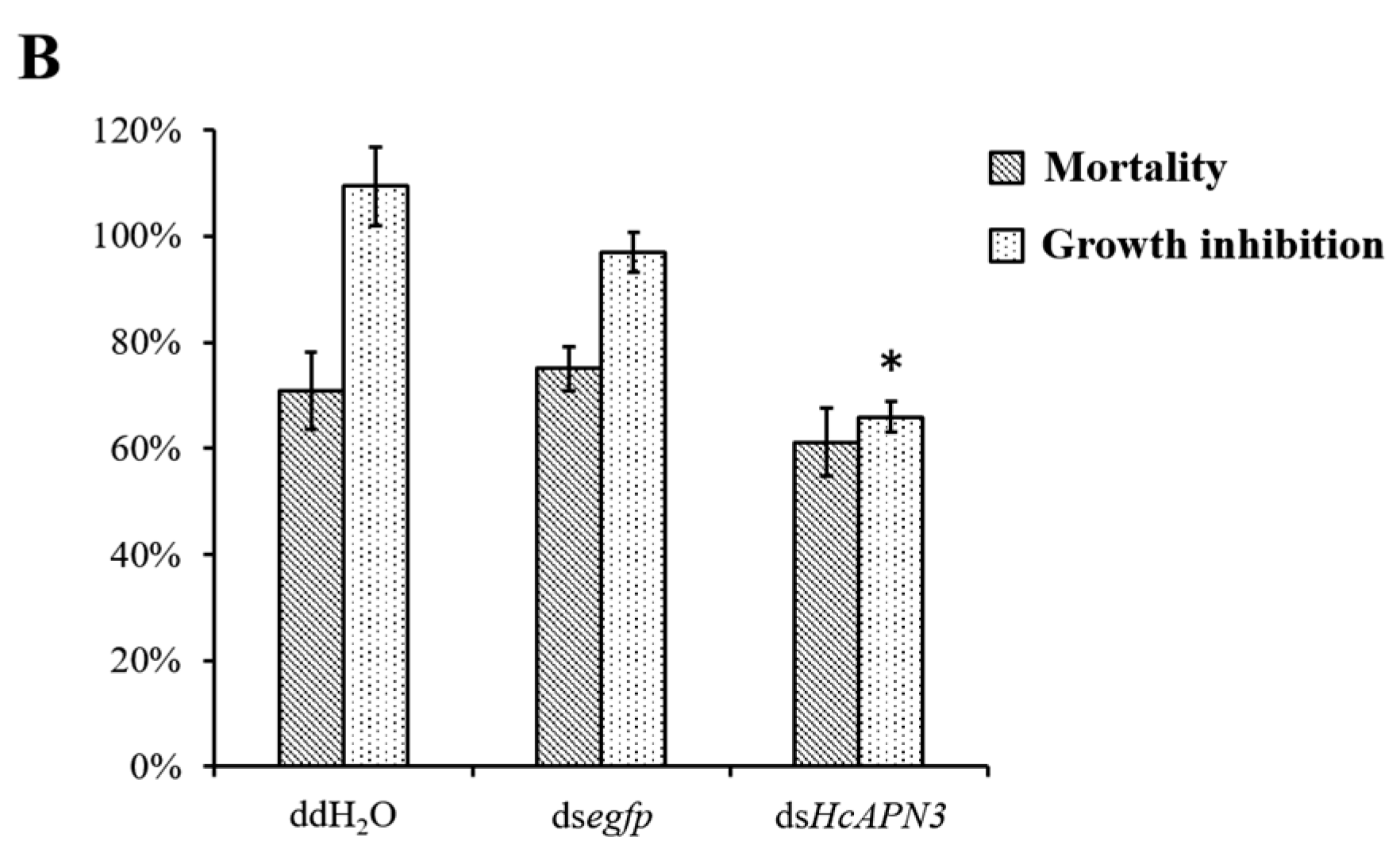
2.7. Silencing of HcAPN3 Expression by RNAi
2.8. Enhancement of Cry1Ab35 Toxicity by HcAPN3G and HcAPN3E
3. Discussion
4. Materials and Methods
4.1. Insects
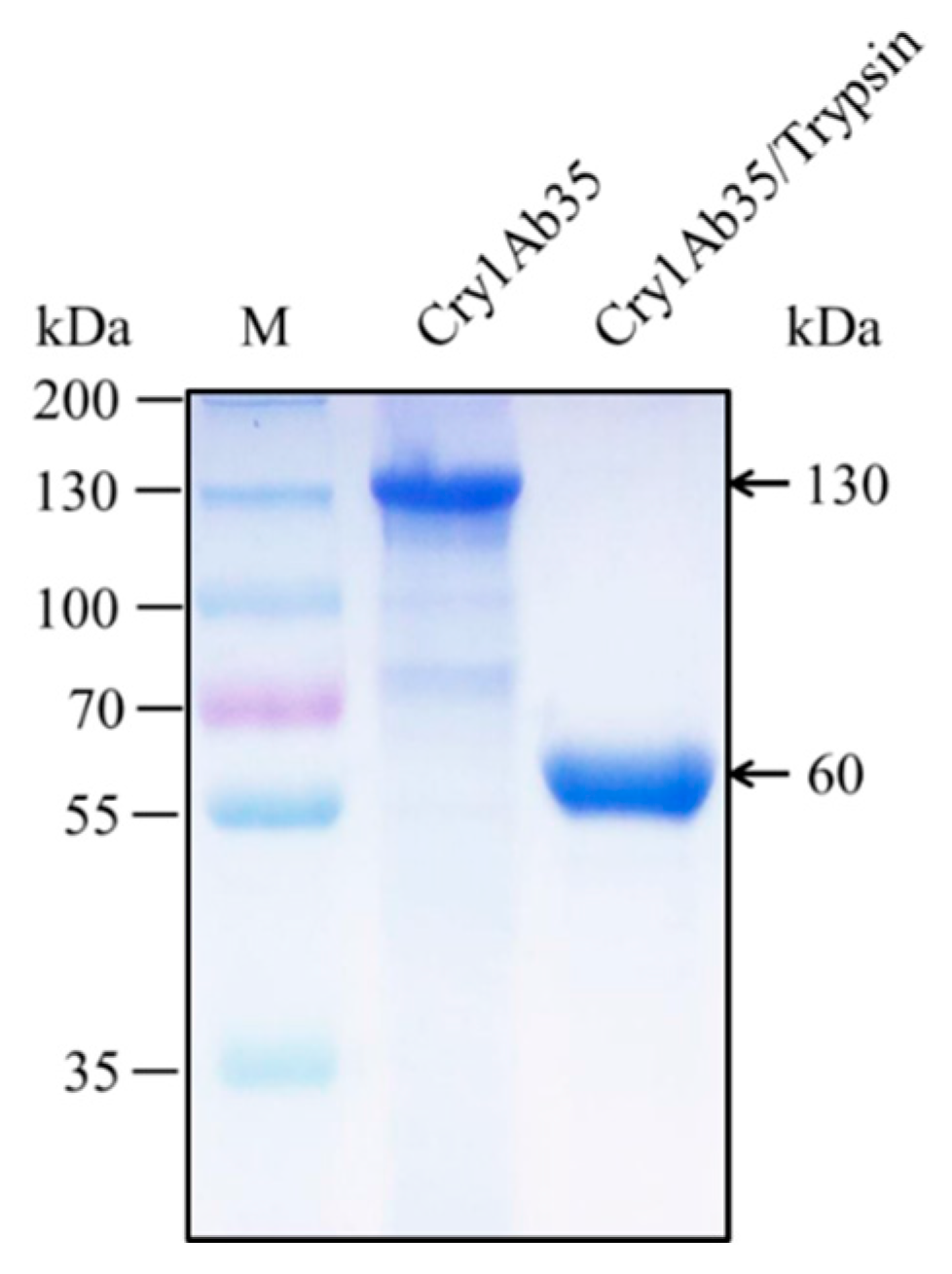
4.2. Preparation of Cry1Ab35 Toxin and Antibody Preparation
4.3. Insect Bioassays
4.4. Preparation of Brush Border Membrane Vesicles (BBMV)
4.5. Ligand Blot and Protein Identification by LC-MS/MS
4.6. Cloning and Sequencing of HcAPN3 from the Midgut of H. cunea Larvae
4.7. Expression of HcAPN3 Fragment Peptides in E. coli
4.8. Expression of HcAPN3 in Sf9 Cells Using a Baculovirus Expression System
4.9. Detection of HcAPN3 Expression and Cry1Ab35 Binding
4.10. Developmental and Tissue Expression Analysis by Semi-Quantitative PCR
4.11. Induced Expression Analysis Using Quantitative Real-Time PCR (qPCR)
4.12. RNA Interference (RNAi)
4.13. Enhancement of HcAPN3 Fragment Peptides to the Toxicity of Cry1Ab35
4.14. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gomi, T. Seasonal adaptations of the fall webworm Hyphantria cunea (Drury) (Lepidoptera: Arctiidae) following its invasion of Japan. Ecol. Res. 2007, 22, 855–861. [Google Scholar] [CrossRef]
- Zhang, L.W.; Kang, K.; Jiang, S.C.; Zhang, Y.N.; Wang, T.T.; Zhang, J.; Sun, L.; Yang, Y.Q.; Huang, C.C.; Jiang, L.Y.; et al. Analysis of the Antennal Transcriptome and Insights into Olfactory Genes in Hyphantria cunea (Drury). PLoS ONE 2016, 11, e0164729. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wei, X.; Xiao, H.; He, H.; Xia, Q.; Xue, F. Diapause induction and termination in Hyphantria cunea (Drury) (Lepidoptera: Arctiinae). PLoS ONE 2014, 9, e98145. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.W.; Kang, K.; Liu, Y.J.; Zhang, J.; Sun, L.; Zhang, C.; Huang, C.C.; Jiang, L.Y.; Ye, K.Y.; Ding, D.G. Evaluation of Beauveria bassiana isolates as potential agents for control of Hyphantria cunea (Lepidoptera: Arctiidae). Acta Entomol. Sin. 2016, 59, 111–118. [Google Scholar]
- Yang, Z.Q.; Wei, J.R.; Wang, X.Y. Mass rearing and augmentative releases of the native parasitoid Chouioia cunea for biological control of the introduced fall webworm Hyphantria cunea in China. BioControl 2006, 51, 401–408. [Google Scholar] [CrossRef]
- Tang, R.; Zhang, J.P.; Zhang, Z.N. Electrophysiological and behavioral responses of male fall webworm moths (Hyphantria cunea) to Herbivory-induced mulberry (Morus alba) leaf volatiles. PLoS ONE 2012, 7, e49256. [Google Scholar] [CrossRef] [PubMed]
- Pigott, C.R.; Ellar, D.J. Role of receptors in Bacillus thuringiensis crystal toxin activity. Microbiol. Mol. Biol. Rev. 2007, 71, 255–281. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Sanchez, J.; Kouskoura, T.; Crickmore, N. N-terminal activation is an essential early step in the mechanism of action of the Bacillus thuringiensis Cry1Ac insecticidal toxin. J. Biol. Chem. 2002, 277, 23985–23987. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Cheng, T.; Jin, S.; Jiang, L.; Wang, C.; Xia, Q. Structural, evolutionary and functional analysis of APN genes in the Lepidoptera Bombyx mori. Gene 2014, 535, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, X.; Zhang, J. Molecular characterization of four midgut aminopeptidase N isozymes from the cabbage looper, Trichoplusia ni. Insect Biochem. Mol. Biol. 2005, 35, 611–620. [Google Scholar] [CrossRef] [PubMed]
- Hooper, N.M. Families of zinc metalloproteases. FEBS Lett. 1994, 354, 1–6. [Google Scholar] [CrossRef]
- Crava, C.M.; Bel, Y.; Lee, S.F.; Manachini, B.; Heckel, D.G.; Escriche, B. Study of the aminopeptidase N gene family in the lepidopterans Ostrinia nubilalis (Hubner) and Bombyx mori (L.): Sequences, mapping and expression. Insect Biochem. Mol. Biol. 2010, 40, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.L. Evolutionary diversification of aminopeptidase N in Lepidoptera by conserved clade-specific amino acid residues. Mol. Phylogen. Evol. 2014, 76, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Du, L.X.; Liu, C.X.; Gong, L.; Han, L.Z.; Peng, Y.F. RNAi in the striped stem borer, Chilo suppressalis, establishes a functional role for aminopeptidase N in Cry1Ab intoxication. J. Invertebr. Pathol. 2016, 143, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bravo, A.; Gill, S.S.; Soberon, M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 2007, 49, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Gomez, I.; Sanchez, J.; Miranda, R.; Bravo, A.; Soberon, M. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 2002, 513, 242–246. [Google Scholar] [CrossRef]
- Arenas, I.; Bravo, A.; Soberon, M.; Gomez, I. Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J. Biol. Chem. 2010, 285, 12497–12503. [Google Scholar] [CrossRef] [PubMed]
- Bretschneider, A.; Heckel, D.G.; Pauchet, Y. Three toxins, two receptors, one mechanism: Mode of action of Cry1A toxins from Bacillus thuringiensis in Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 76, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Miyamoto, K.; Yamamoto, K.; Narukawa, J.; Kawai, S.; Sezutsu, H.; Kobayashi, I.; Uchino, K.; Tamura, T.; Mita, K.; et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc. Natl. Acad. Sci. USA 2012, 109, E1591–E1598. [Google Scholar] [CrossRef] [PubMed]
- Knight, P.J.; Crickmore, N.; Ellar, D.J. The receptor for Bacillus thuringiensis CrylA(c) delta-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 1994, 11, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, C.; Barrett-Wilt, G.A.; Hunt, D.F.; Akhurst, R.J.; East, P.D.; Gordon, K.H.; Campbell, P.M. Diversity of aminopeptidases, derived from four lepidopteran gene duplications, and polycalins expressed in the midgut of Helicoverpa armigera: Identification of proteins binding the delta-endotoxin, Cry1Ac of Bacillus thuringiensis. Insect Biochem. Mol. Biol. 2008, 38, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Crava, C.M.; Bel, Y.; Jakubowska, A.K.; Ferre, J.; Escriche, B. Midgut aminopeptidase N isoforms from Ostrinia nubilalis: Activity characterization and differential binding to Cry1Ab and Cry1Fa proteins from Bacillus thuringiensis. Insect Biochem. Mol. Biol. 2013, 43, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Herrero, S.; Gechev, T.; Bakker, P.L.; Moar, W.J.; de Maagd, R.A. Bacillus thuringiensis Cry1Ca-resistant Spodoptera exigua lacks expression of one of four Aminopeptidase N genes. BMC Genom. 2005, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cheng, H.; Gao, Y.; Wang, G.; Liang, G.; Wu, K. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem. Mol. Biol. 2009, 39, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Tiewsiri, K.; Wang, P. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. USA 2011, 108, 14037–14042. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, B.D.; Hellmich, R.L. Understanding successful resistance management: The European corn borer and Bt corn in the United States. GM Crops Food 2012, 3, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tian, J.C.; Shen, Z.C.; Peng, Y.F.; Hu, C.; Guo, Y.Y.; Ye, G.Y. Transgenic rice plants expressing a fused protein of Cry1Ab/Vip3H has resistance to rice stem borers under laboratory and field conditions. J. Econ. Entomol. 2010, 103, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Ibiza-Palacios, M.S.; Ferre, J.; Higurashi, S.; Miyamoto, K.; Sato, R.; Escriche, B. Selective inhibition of binding of Bacillus thuringiensis Cry1Ab toxin to cadherin-like and aminopeptidase proteins in brush-border membranes and dissociated epithelial cells from Bombyx mori. Biochem. J. 2008, 409, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Miyamoto, K.; Noda, H.; Endo, H.; Kikuta, S.; Sato, R. Single amino acid insertions in extracellular loop 2 of Bombyx mori ABCC2 disrupt its receptor function for Bacillus thuringiensis Cry1Ab and Cry1Ac but not Cry1Aa toxins. Peptides 2016, 78, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Flores-Escobar, B.; Rodriguez-Magadan, H.; Bravo, A.; Soberon, M.; Gomez, I. Differential role of Manduca sexta aminopeptidase-N and alkaline phosphatase in the mode of action of Cry1Aa, Cry1Ab, and Cry1Ac toxins from Bacillus thuringiensis. Appl. Environ. Microbiol. 2013, 79, 4543–4550. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.D.; Yu, C.G.; Mathis, J.P.; Meyer, T.E.; Shi, X.; Siqueira, H.A.; Siegfried, B.D. Identification, cloning and expression of a Cry1Ab cadherin receptor from European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 2005, 35, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Ferre, J.; Van Rie, J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 2002, 47, 501–533. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.E.; Young, M.K.; Lee, T.D. Qscore: An algorithm for evaluating SEQUEST database search results. J. Am. Soc. Mass Spectrom. 2002, 13, 378–386. [Google Scholar] [CrossRef]
- Luo, K.; Banks, D.; Adang, M.J. Toxicity, binding, and permeability analyses of four Bacillus thuringiensis Cry1 delta-endotoxins using brush border membrane vesicles of Spodoptera exigua and Spodoptera frugiperda. Appl. Environ. Microbiol. 1999, 65, 457–464. [Google Scholar] [PubMed]
- Wolfersberger, M.G. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the gypsy moth (Lymantria dispar). Arch. Insect Biochem. Physiol. 1993, 24, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Wolfersberger, M.; Luethy, P.; Maurer, A.; Parenti, P.; Sacchi, F.V.; Glordana, B.; Hanozet, G.M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. 1987, 86, 301–308. [Google Scholar] [CrossRef]
- Eisenhaber, B.; Bork, P.; Eisenhaber, F. Post-translational GPI lipid anchor modification of proteins in kingdoms of life: Analysis of protein sequence data from complete genomes. Protein Eng. 2001, 14, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Hoogland, C.; Mostaguir, K.; Appel, R.D.; Lisacek, F. The World-2DPAGE Constellation to promote and publish gel-based proteomics data through the ExPASy server. J. Proteom. 2008, 71, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Banks, D.J.; Jurat-Fuentes, J.L.; Dean, D.H.; Adang, M.J. Bacillus thuringiensis Cry1Ac and Cry1Fa delta-endotoxin binding to a novel 110 kDa aminopeptidase in Heliothis virescens is not N-acetylgalactosamine mediated. Insect Biochem. Mol. Biol. 2001, 31, 909–918. [Google Scholar] [CrossRef]
- Ning, C.; Wu, K.; Liu, C.; Gao, Y.; Jurat-Fuentes, J.L.; Gao, X. Characterization of a Cry1Ac toxin-binding alkaline phosphatase in the midgut from Helicoverpa armigera (Hubner) larvae. J. Insect Physiol. 2010, 56, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Rajagopal, R.; Sivakumar, S.; Agrawal, N.; Malhotra, P.; Bhatnagar, R.K. Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem. 2002, 277, 46849–46851. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; Rajagopal, R.; Venkatesh, G.R.; Srivastava, A.; Bhatnagar, R.K. Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J. Biol. Chem. 2007, 282, 7312–7319. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Chen, R.R.; Zhang, Y.; Ma, Y.; Cui, J.J.; Han, Z.J.; Mu, L.L.; Li, G.Q. A Spodoptera exigua cadherin serves as a putative receptor for Bacillus thuringiensis Cry1Ca toxin and shows differential enhancement of Cry1Ca and Cry1Ac toxicity. Appl. Environ. Microbiol. 2013, 79, 5576–5583. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Zhao, M.; Wei, J.; Zhang, W.; Wang, B.; Myint Khaing, M.; Liang, G. New insights on the role of alkaline phosphatase 2 from Spodoptera exigua (Hubner) in the action mechanism of Bt toxin Cry2Aa. J. Insect Physiol. 2017, 98, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhu, Y.C.; Ottea, J.; Husseneder, C.; Leonard, B.R.; Abel, C.; Huang, F. Molecular characterization and RNA interference of three midgut aminopeptidase N isozymes from Bacillus thuringiensis-susceptible and -resistant strains of sugarcane borer, Diatraea saccharalis. Insect Biochem. Mol. Biol. 2010, 40, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Ma, Y.; Cui, J.J.; Li, G.Q. RNA interference-mediated knockdown of three putative aminopeptidases N affects susceptibility of Spodoptera exigua larvae to Bacillus thuringiensis Cry1Ca. J. Insect Physiol. 2014, 67, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Aimanova, K.G.; Fernandez, L.E.; Bravo, A.; Soberon, M.; Gill, S.S. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem. J. 2009, 424, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, A.I.; Reyes, E.Z.; Cancino-Rodezno, A.; Bedoya-Perez, L.P.; Caballero-Flores, G.G.; Muriel-Millan, L.F.; Likitvivatanavong, S.; Gill, S.S.; Bravo, A.; Soberon, M. Aedes aegypti alkaline phosphatase ALP1 is a functional receptor of Bacillus thuringiensis Cry4Ba and Cry11Aa toxins. Insect Biochem. Mol. Biol. 2012, 42, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Aimanova, K.G.; Pan, S.; Gill, S.S. Identification and characterization of Aedes aegypti aminopeptidase N as a putative receptor of Bacillus thuringiensis Cry11A toxin. Insect Biochem. Mol. Biol. 2009, 39, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Likitvivatanavong, S.; Aimanova, K.G.; Gill, S.S. A 104 kDa Aedes aegypti aminopeptidase N is a putative receptor for the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Insect Biochem. Mol. Biol. 2013, 43, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hua, G.; Jurat-Fuentes, J.L.; Abdullah, M.A.; Adang, M.J. Synergism of Bacillus thuringiensis toxins by a fragment of a toxin-binding cadherin. Proc. Natl. Acad. Sci. USA 2007, 104, 13901–13906. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Hua, G.; Urbauer, J.L.; Adang, M.J. Synergistic and inhibitory effects of aminopeptidase peptides on Bacillus thuringiensis Cry11Ba toxicity in the mosquito Anopheles gambiae. Biochemistry 2010, 49, 8512–8519. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Liang, G.M.; Crickmore, N.; Li, H.T.; He, K.L.; Song, F.P.; Feng, X.; Huang, D.F.; Zhang, J. Cloning and characterization of a novel Cry1A toxin from Bacillus thuringiensis with high toxicity to the Asian corn borer and other lepidopteran insects. FEMS Microbiol. Lett. 2008, 280, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]


| Protein | Protein Qscore | Protein Mass (kDa) | Unique Peptide Num | Coverage (%) | Alignment Annotation |
|---|---|---|---|---|---|
| CL698_Contig1_All | 69 | 92.8 | 21 | 32 | heat shock protein-like [B. mori] (XP_004927222.1) |
| CL2518_Contig2_All | 68 | 104.4 | 22 | 25 | aminopeptidase N-10 [B. mori] (AFK85026.1) |
| CL1273_Contig2_All | 41 | 107.8 | 12 | 17 | aminopeptidase N3 [L. dispar] (AAL26894.1) |
| CL2004_Contig1_All | 39 | 111.6 | 13 | 18 | aminopeptidase N3 [Danaus plexippus] (EHJ64337) |
| CL2587_Contig1_All | 25 | 67.3 | 8 | 16 | proton ATPase catalytic subunit A [M. sexta] (P31400.1) |
| Unigene188_All | 22 | 104.5 | 7 | 11 | aminopeptidase N-9 [B. mori] (AFK85025.1) |
| CL593_Contig3_All | 9 | 67.0 | 3 | 8 | aminopeptidase N2 [B. mori] (AFK85018) |
| CL2619_Contig1_All | 8 | 114.2 | 2 | 6 | aminopeptidase N1 [L. dispar] (AAD31183.1) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Zhao, D.; Yan, X.; Guo, W.; Bao, Y.; Wang, W.; Wang, X. Identification and Characterization of Hyphantria cunea Aminopeptidase N as a Binding Protein of Bacillus thuringiensis Cry1Ab35 Toxin. Int. J. Mol. Sci. 2017, 18, 2575. https://doi.org/10.3390/ijms18122575
Zhang Y, Zhao D, Yan X, Guo W, Bao Y, Wang W, Wang X. Identification and Characterization of Hyphantria cunea Aminopeptidase N as a Binding Protein of Bacillus thuringiensis Cry1Ab35 Toxin. International Journal of Molecular Sciences. 2017; 18(12):2575. https://doi.org/10.3390/ijms18122575
Chicago/Turabian StyleZhang, Yakun, Dan Zhao, Xiaoping Yan, Wei Guo, Yajun Bao, Wei Wang, and Xiaoyun Wang. 2017. "Identification and Characterization of Hyphantria cunea Aminopeptidase N as a Binding Protein of Bacillus thuringiensis Cry1Ab35 Toxin" International Journal of Molecular Sciences 18, no. 12: 2575. https://doi.org/10.3390/ijms18122575





