Poly-γ-glutamic Acid Synthesis, Gene Regulation, Phylogenetic Relationships, and Role in Fermentation
Abstract
:1. Introduction
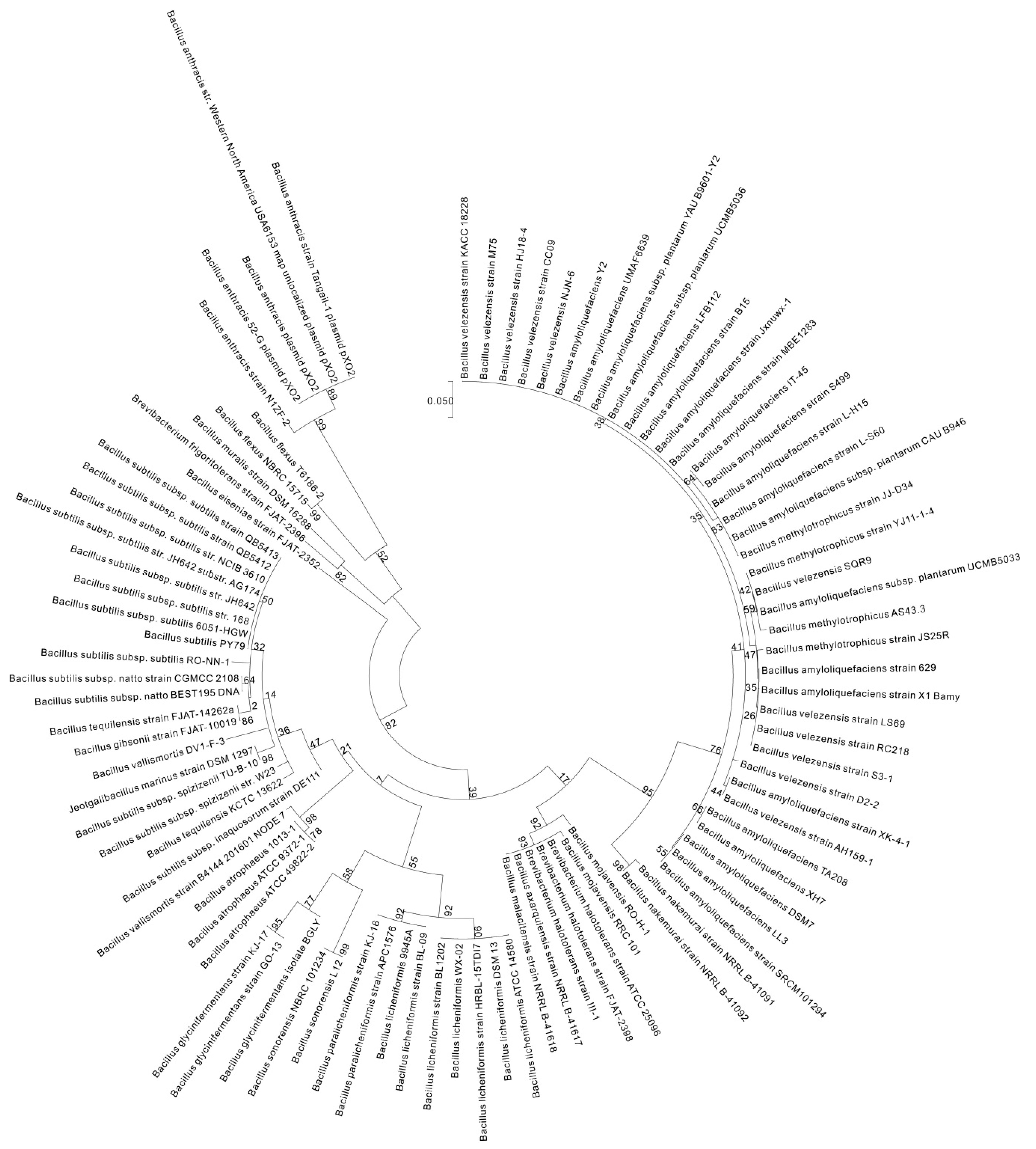
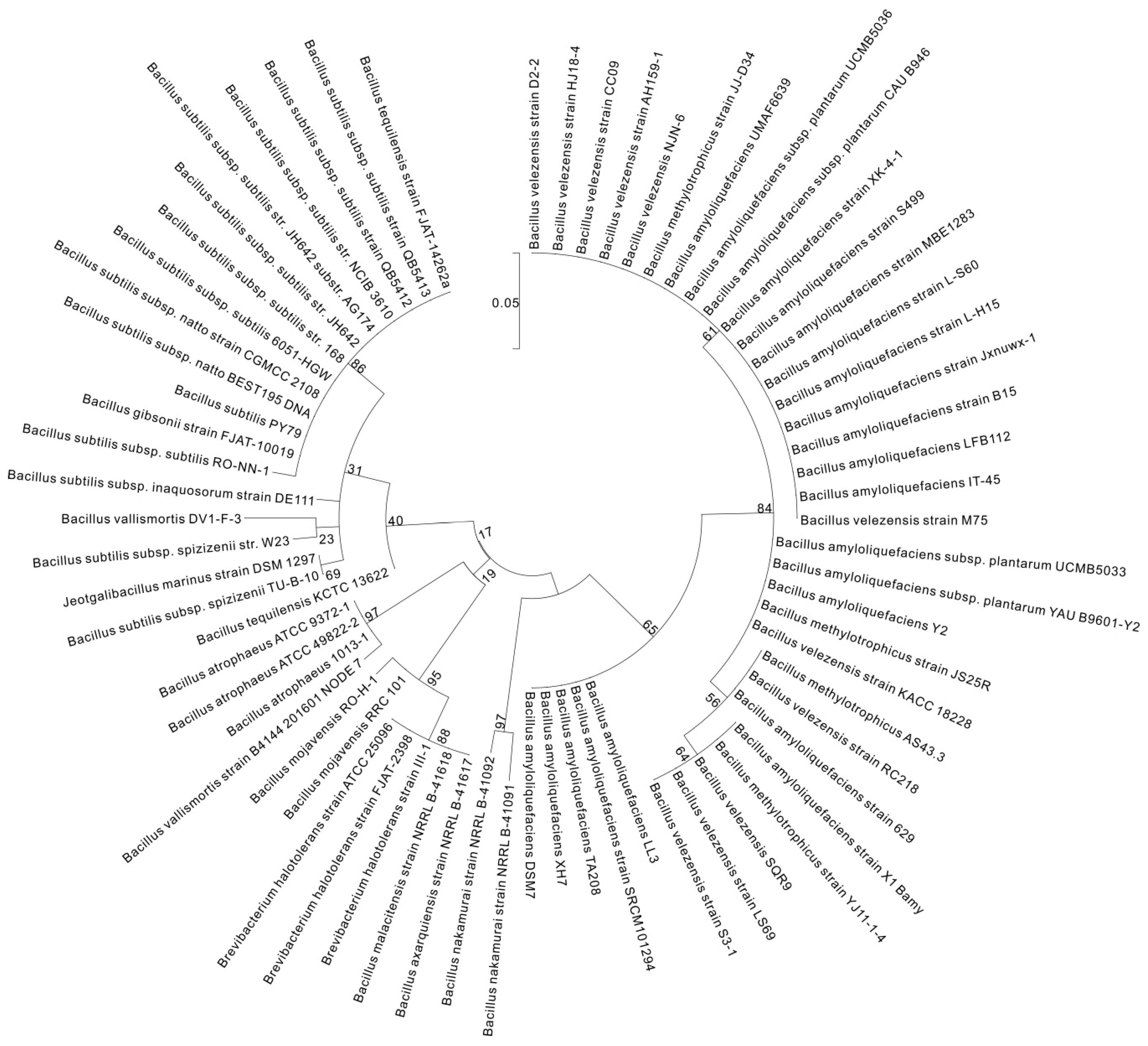
2. γ-PGA Producers
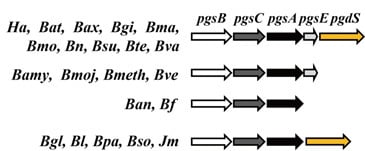
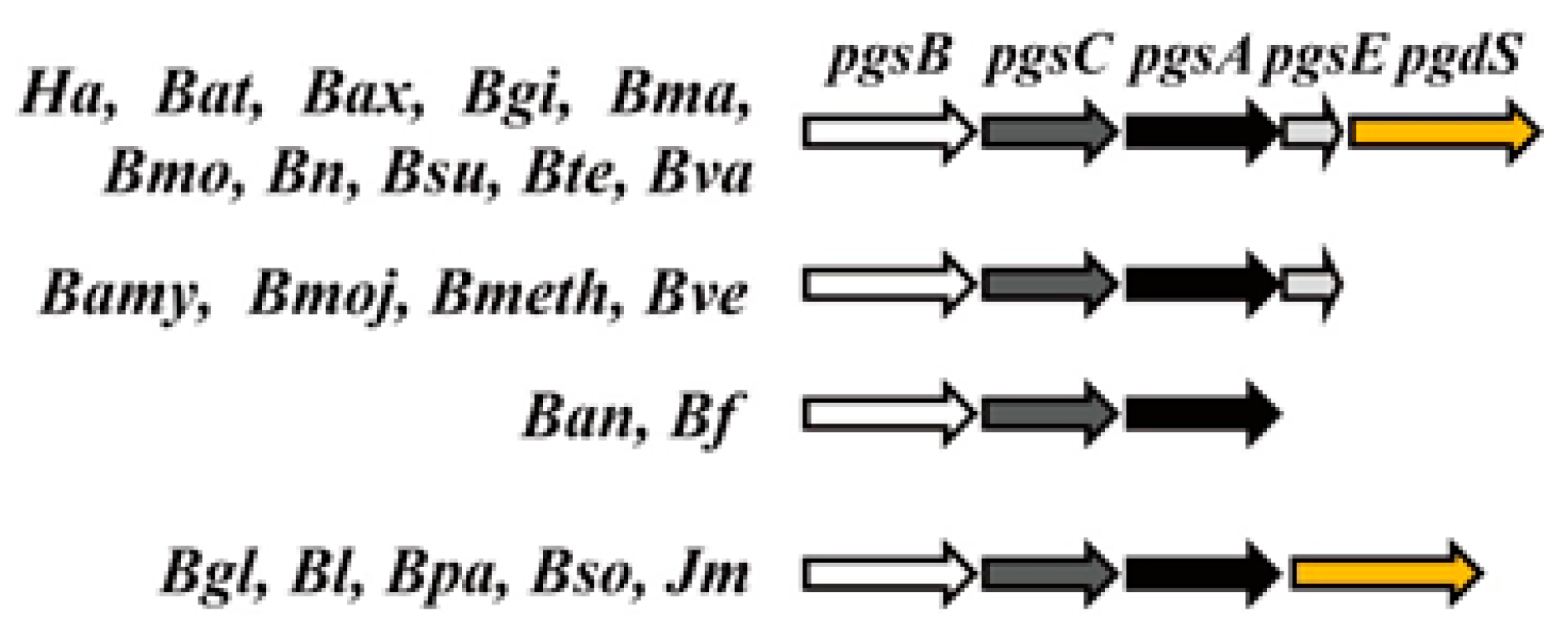
3. Genes Involved in γ-PGA Synthesis and Degradation
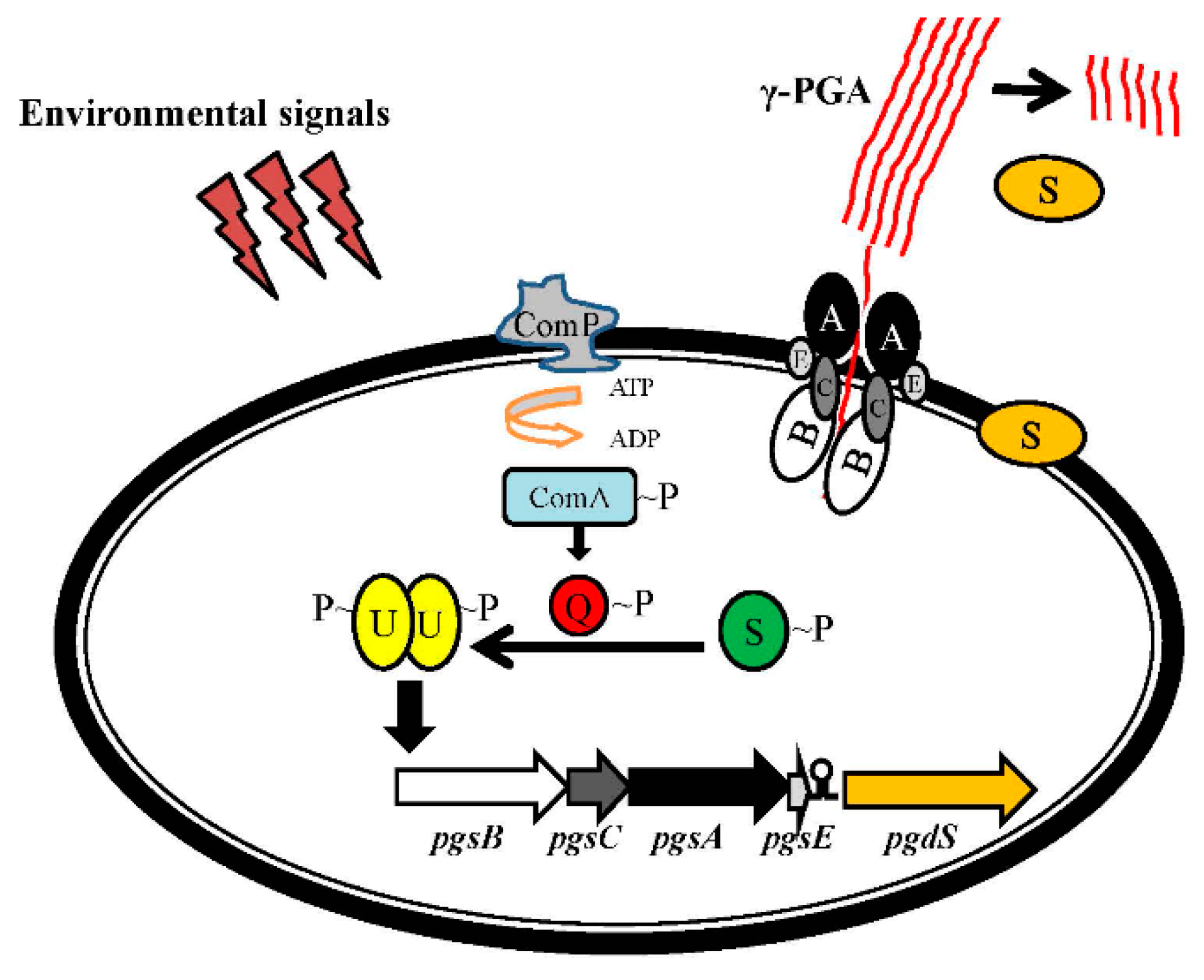
4. Regulation of the pgsBCA Operon
5. Recombinant Strains Used for γ-PGA Production
6. Fermentation Conditions
7. Applications of γ-PGA
8. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| γ-PGA | Poly-γ-glutamic acid |
| TCA | Tricarboxylic acid |
| ATP | Adenosine triphosphate |
| MCL | Maximum composite likelihood |
| CoA | Coenzyme |
| EPS | Exopolysaccharides |
References
- Shi, F.; Xu, Z.; Cen, P. Microbial production of natural poly amino acid. Sci. China Ser. B 2007, 50, 291. [Google Scholar] [CrossRef]
- Ashiuchi, M. Occurrence and biosynthetic mechanism of poly-γ-glutamic acid. In Amino-Acid Homopolymers Occurring in Nature; Springer: Berlin, Germany, 2010; pp. 77–93. [Google Scholar]
- Otani, Y.; Tabata, Y.; Ikada, Y. A new biological glue from gelatin and poly (l-glutamic acid). J. Biomed. Mater. Res. 1996, 31, 157–166. [Google Scholar] [CrossRef]
- Otani, Y.; Tabata, Y.; Ikada, Y. Sealing effect of rapidly curable gelatin-poly (l-glutamic acid) hydrogel glue on lung air leak. Ann. Thorac. Surg. 1999, 67, 922–926. [Google Scholar] [CrossRef]
- Kurosaki, T.; Kitahara, T.; Fumoto, S.; Nishida, K.; Nakamura, J.; Niidome, T.; Kodama, Y.; Nakagawa, H.; To, H.; Sasaki, H. Ternary complexes of pDNA, polyethylenimine, and γ-polyglutamic acid for gene delivery systems. Biomaterials 2009, 30, 2846–2853. [Google Scholar] [CrossRef] [PubMed]
- Shyu, Y.S.; Sung, W.C. Improving the emulsion stability of sponge cake by the addition of γ-polyglutamic acid. J. Mar. Sci. Technol. 2010, 18, 895–900. [Google Scholar]
- Shyu, Y.S.; Hwang, J.Y.; Hsu, C.K. Improving the rheological and thermal properties of wheat dough by the addition of γ-polyglutamic acid. LWT Food. Sci. Technol. 2008, 41, 982–987. [Google Scholar] [CrossRef]
- Lim, S.M.; Kim, J.; Shim, J.Y.; Imm, B.Y.; Sung, M.H.; Imm, J.Y. Effect of poly-γ-glutamic acids (PGA) on oil uptake and sensory quality in doughnuts. Food. Sci. Biotechnol. 2012, 21, 247–252. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Hestekin, J.; Brushaber, P.; Cullen, L.; Bachas, L.; Sikdar, S. Novel poly-glutamic acid functionalized microfiltration membranes for sorption of heavy metals at high capacity. J. Membr. Sci. 1998, 141, 121–135. [Google Scholar] [CrossRef]
- Inbaraj, B.S.; Chen, B.H. In vitro removal of toxic heavy metals by poly(γ-glutamic acid)-coated superparamagnetic nanoparticles. Int. J. Nanomed. 2012, 7, 4419–4432. [Google Scholar]
- Hanby, W.; Rydon, H. The capsular substance of Bacillus anthracis: With an appendix by P. Bruce White. Biochem. J. 1946, 40, 297. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Kamei, T.; Baek, D.H.; Shin, S.Y.; Sung, M.H.; Soda, K.; Yagi, T.; Misono, H. Isolation of Bacillus subtilis (chungkookjang), a poly-γ-glutamate producer with high genetic competence. Appl. Microbiol. Biotechnol. 2001, 57, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Ueda, S. Regulation of poly glutamate production in Bacillus subtilis (natto): Transformation of high PGA productivity. Agric. Biol. Chem. 1982, 46, 2275–2281. [Google Scholar]
- Zhao, C.; Zhang, Y.; Wei, X.; Hu, Z.; Zhu, F.; Xu, L.; Luo, M.; Liu, H. Production of ultra-high molecular weight poly-γ-glutamic acid with Bacillus licheniformis P-104 and characterization of its flocculation properties. Appl. Biochem. Biotechnol. 2013, 170, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Tran, L.S.P.; Uchida, I.; Itoh, Y. Characterization of Bacillus subtilis γ-glutamyltransferase and its involvement in the degradation of capsule poly-γ-glutamate. Microbiology 2004, 150, 4115–4123. [Google Scholar] [CrossRef] [PubMed]
- Lazazzera, B. Cell-density control of gene expression and development in Bacillus subtilis. Cell Cell Signal. Bact. 1999, 1999, 27–47. [Google Scholar]
- Bajaj, I.B.; Singhal, R.S. Enhanced production of poly(γ-glutamic acid) from Bacillus licheniformis NCIM 2324 by using metabolic precursors. Appl. Biochem. Biotechnol. 2009, 159, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Soda, K.; Misono, H. Characterization of yrpc gene product of Bacillus subtilis IFO 3336 as glutamate racemase isozyme. Biosci. Biotechnol. Biochem. 1999, 63, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Kunioka, M. Biosynthesis and hydrolysis of poly(γ-glutamic acid) from Bacillus subtilis IF03335. Biosci. Biotechnol. Biochem. 1992, 56, 1031–1035. [Google Scholar] [CrossRef] [PubMed]
- Shih, L.; Wu, P.J.; Shieh, C.J. Microbial production of a poly(γ-glutamic acid) derivative by Bacillus subtilis. Process Biochem. 2005, 40, 2827–2832. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Nawa, C.; Kamei, T.; Song, J.J.; Hong, S.P.; Sung, M.H.; Soda, K.; Yagi, T.; Misono, H. Physiological and biochemical characteristics of poly γ-glutamate synthetase complex of Bacillus subtilis. FEBS J. 2001, 268, 5321–5328. [Google Scholar]
- Troy, F.A. Chemistry and biosynthesis of the poly(γ-d-glutamyl) capsule in Bacillus licheniformis I. Properties of the membrane-mediated biosynthetic reaction. J. Biol. Chem. 1973, 248, 305–315. [Google Scholar] [PubMed]
- Kocianova, S.; Vuong, C.; Yao, Y.; Voyich, J.M.; Fischer, E.R.; DeLeo, F.R.; Otto, M. Key role of poly-γ-dl-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Investig. 2005, 115, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Hezayen, F.; Rehm, B.; Tindall, B.; Steinbüchel, A. Transfer of Natrialba asiatica B1T to Natrialba taiwanensis sp. nov. and description of Natrialba aegyptiaca sp. nov., a novel extremely halophilic, aerobic, non-pigmented member of the archaea from Egypt that produces extracellular poly(glutamic acid). Int. J. Syst. Evol. Microbiol. 2001, 51, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Cromwick, A.M.; Gross, R.A. Effects of Manganese (II) on Bacillus licheniformis ATCC 9945A physiology and γ-poly(glutamic acid) formation. Int. J. Biol. Macromol. 1995, 17, 259–267. [Google Scholar] [CrossRef]
- Thorne, C.B.; Leonard, C.G. Isolation of d-and l-glutamyl polypeptides from culture filtrates of Bacillus subtilis. J. Biol. Chem. 1958, 233, 1109–1112. [Google Scholar] [PubMed]
- Shimizu, K.; Nakamura, H.; Ashiuchi, M. Salt-inducible bionylon polymer from Bacillus megaterium. Appl. Environ. Microbiol. 2007, 73, 2378–2379. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Kamei, T.; Misono, H. Poly-γ-glutamate synthetase of Bacillus subtilis. J. Mol. Catal. B Enzym. 2003, 23, 101–106. [Google Scholar] [CrossRef]
- Choi, S.H.; Park, J.S.; Yoon, K.; Choi, W. Production of microbial biopolymer, poly(γ-glutamic acid) by Bacillus subtilis BS 62. Agric. Chem. Biotechnol. 2004, 47, 60–64. [Google Scholar]
- Ashiuchi, M.; Soda, K.; Misono, H. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO 3336: Gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 1999, 263, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Candela, T.; Mock, M.; Fouet, A. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J. Bacteriol. 2005, 187, 7765–7772. [Google Scholar] [CrossRef] [PubMed]
- Tarui, Y.; Iida, H.; Ono, E.; Miki, W.; Hirasawa, E.; Fujita, K.I.; Tanaka, T.; Taniguchi, M. Biosynthesis of poly-γ-glutamic acid in plants: Transient expression of poly-γ-glutamate synthetase complex in tobacco leaves. J. Biosci. Bioeng. 2005, 100, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Shang, L.; Yoon, S.H.; Lee, S.Y.; Yu, Z. Optimal production of poly-γ-glutamic acid by metabolically engineered Escherichia coli. Biotechnol. Lett. 2006, 28, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Drysdale, M.; Bourgogne, A.; Koehler, T.M. Transcriptional analysis of the Bacillus anthracis capsule regulators. J. Bacteriol. 2005, 187, 5108–5114. [Google Scholar] [CrossRef] [PubMed]
- Leonard, C.G.; Housewright, R.D.; Thorne, C.B. Effects of some metallic ions on glutamyl polypeptide synthesis by Bacillus subtilis. J. Bacteriol. 1958, 76, 499. [Google Scholar] [PubMed]
- Suzuki, T.; Tahara, Y. Characterization of the Bacillus subtilis ywtD gene, whose product is involved in γ-polyglutamic acid degradation. J. Bacteriol. 2003, 185, 2379–2382. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.P.; Nagai, T.; Itoh, Y. Divergent structure of the ComQXPA quorum-sensing components: Molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 2000, 37, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Stanley, N.R.; Lazazzera, B.A. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-γ-DL-glutamic acid production and biofilm formation. Mol. Microbiol. 2005, 57, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Osera, C.; Amati, G.; Calvio, C.; Galizzi, A. SwrAA activates poly-γ-glutamate synthesis in addition to swarming in Bacillus subtilis. Microbiology 2009, 155, 2282–2287. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.; Suzuki, Y.; Abe, N.; Kaneko, J.; Itoh, Y.; Kimura, K. Mutations suppressing the loss of DegQ function in Bacillus subtilis (natto) poly-γ-glutamate synthesis. Appl. Environ. Microbiol. 2011, 77, 8249–8258. [Google Scholar] [CrossRef] [PubMed]
- Cairns, L.S.; Marlow, V.L.; Bissett, E.; Ostrowski, A.; Stanley-Wall, N.R. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol. Microbiol. 2013, 90, 6–21. [Google Scholar] [PubMed]
- Feng, J.; Gao, W.; Gu, Y.; Zhang, W.; Cao, M.; Song, C.; Zhang, P.; Sun, M.; Yang, C.; Wang, S. Functions of poly-γ-glutamic acid (γ-PGA) degradation genes in γ-PGA synthesis and cell morphology maintenance. Appl. Microbiol. Biotechnol. 2014, 98, 6397–6407. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Gu, Y.; Sun, Y.; Han, L.; Yang, C.; Zhang, W.; Cao, M.; Song, C.; Gao, W.; Wang, S. Metabolic engineering of Bacillus amyloliquefaciens for poly-γ-glutamic acid (γ-PGA) overproduction. Microb. Biotechnol. 2014, 7, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; He, Y.; Gao, W.; Feng, J.; Cao, M.; Yang, C.; Song, C.; Wang, S. Deletion of genes involved in glutamate metabolism to improve poly-γ-glutamic acid production in B. amyloliquefaciens LL3. J. Ind. Microbiol. Biotechnol. 2015, 42, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Scoffone, V.; Dondi, D.; Biino, G.; Borghese, G.; Pasini, D.; Galizzi, A.; Calvio, C. Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis. Biotechnol. Bioeng. 2013, 110, 2006–2012. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Li, Z.; Zhang, H.; Wu, J.; Luo, M. Cloning and expression of the γ-polyglutamic acid synthetase gene pgsBCA in Bacillus subtilis WB600. BioMed Res. Int. 2016, 2016, 3073949. [Google Scholar] [PubMed]
- Yamashiro, D.; Yoshioka, M.; Ashiuchi, M. Bacillus subtilis pgsE (formerly ywtC) stimulates poly-γ-glutamate production in the presence of zinc. Biotechnol. Bioeng. 2011, 108, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Qi, G.; Ji, Z.; Zhang, S.; Liu, J.; Ma, X.; Chen, S. Expression of glr gene encoding glutamate racemase in Bacillus licheniformis WX-02 and its regulatory effects on synthesis of poly-γ-glutamic acid. Biotechnol. Lett. 2011, 33, 1837. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Fu, J.; Wei, X.; Ji, Z.; Ma, X.; Qi, G.; Chen, S. Enhanced expression of pgdS gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J. Chem. Technol. Biotechnol. 2014, 89, 1825–1832. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Shimanouchi, K.; Horiuchi, T.; Kamei, T.; Misono, H. Genetically engineered poly-γ-glutamate producer from Bacillus subtilis ISW1214. Biosci. Biotechnol. Biochem. 2006, 70, 1794–1797. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Gu, Y.; Quan, Y.; Cao, M.; Gao, W.; Zhang, W.; Wang, S.; Yang, C.; Song, C. Improved poly-γ-glutamic acid production in Bacillus amyloliquefaciens by modular pathway engineering. Metab. Eng. 2015, 32, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.J.; Blackman, S.A.; Foster, S.J. Autolysins of Bacillus subtilis: Multiple enzymes with multiple functions. Microbiology 2000, 146, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Carlsson, P.; Hederstedt, L. Genetic characterization of Bacillus subtilis odhA and odhB, encoding 2-oxoglutarate dehydrogenase and dihydrolipoamide transsuccinylase, respectively. J. Bacteriol. 1989, 171, 3667–3672. [Google Scholar] [CrossRef] [PubMed]
- Commichau, F.M.; Herzberg, C.; Tripal, P.; Valerius, O.; Stülke, J. A regulatory protein–protein interaction governs glutamate biosynthesis in Bacillus subtilis: The glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol. Microbiol. 2007, 65, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; González-Pastor, J.E.; Dervyn, E.; Ehrlich, S.D.; Losick, R.; Kolter, R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 2004, 186, 3970–3979. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; Chu, F.; Kearns, D.B.; Losick, R.; Kolter, R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2006, 59, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Ashiuchi, M.; Tani, K.; Soda, K.; Misono, H. Properties of glutamate racemase from Bacillus subtilis IFO 3336 producing poly-γ-glutamate. J. Biochem. 1998, 123, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Du, Y.; Xu, G.; Zhang, H.; Zhu, F.; Huang, L.; Xu, Z. High yield and cost-effective production of poly(γ-glutamic acid) with Bacillus subtilis. Eng. Life Sci. 2011, 11, 291–297. [Google Scholar] [CrossRef]
- Tang, B.; Lei, P.; Xu, Z.; Jiang, Y.; Xu, Z.; Liang, J.; Feng, X.; Xu, H. Highly efficient rice straw utilization for poly-(γ-glutamic acid) production by Bacillus subtilis NX-2. Bioresour. Technol. 2015, 193, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Xu, H.; Liang, J.; Yao, J. Contribution of glycerol on production of poly(γ-glutamic acid) in Bacillus subtilis NX-2. Appl. Biochem. Biotechnol. 2010, 160, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, I.B.; Lele, S.S.; Singhal, R.S. Enhanced production of poly(γ-glutamic acid) from Bacillus licheniformis NCIM 2324 in solid state fermentation. J. Ind. Microbiol. Biotechnol. 2008, 35, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Konglom, N.; Chuensangjun, C.; Pechyen, C.; Sirisansaneeyakul, S. Production of poly-γ-glutamic acid by Bacillus licheniformis: Synthesis and characterization. J. Met. Mater. Miner. 2012, 22, 7–11. [Google Scholar]
- Shi, F.; Xu, Z.; Cen, P. Optimization of γ-polyglutamic acid production by Bacillus subtilis ZJU-7 using a surface-response methodology. Biotechnol. Bioprocess Eng. 2006, 11, 251–257. [Google Scholar] [CrossRef]
- Zhu, F.; Cai, J.; Zheng, Q.; Zhu, X.; Cen, P.; Xu, Z. A novel approach for poly-γ-glutamic acid production using xylose and corncob fibres hydrolysate in Bacillus subtillis HB-1. J. Chem. Technol. Biotechnol. 2014, 89, 616–622. [Google Scholar] [CrossRef]
- Mabrouk, M.; Abou-Zeid, D.; Sabra, W. Application of plackett–burman experimental design to evaluate nutritional requirements for poly(γ-glutamic acid) production in batch fermentation by Bacillus licheniformis A13. Afr. J. Appl. Microbiol. Res. 2012, 1, 6–18. [Google Scholar]
- Kongklom, N.; Luo, H.; Shi, Z.; Pechyen, C.; Chisti, Y.; Sirisansaneeyakul, S. Production of poly-γ-glutamic acid by glutamic acid-independent Bacillus licheniformis TISTR 1010 using different feeding strategies. Biochem. Eng. J. 2015, 100, 67–75. [Google Scholar] [CrossRef]
- Xu, Z.; Feng, X.; Zhang, D.; Tang, B.; Lei, P.; Liang, J.; Xu, H. Enhanced poly(γ-glutamic acid) fermentation by Bacillus subtilis NX-2 immobilized in an aerobic plant fibrous-bed bioreactor. Bioresour. Technol. 2014, 155, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, H.; Shi, N.; Cao, X.; Feng, X.; Li, S.; Ouyang, P. Analysis of carbon metabolism and improvement of γ-polyglutamic acid production from Bacillus subtilis NX-2. Appl. Biochem. Biotechnol. 2010, 160, 2332–2341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Xu, Z.; Xu, H.; Feng, X.; Li, S.; Cai, H.; Wei, Y.; Ouyang, P. Improvement of poly(γ-glutamic acid) biosynthesis and quantitative metabolic flux analysis of a two-stage strategy for agitation speed control in the culture of Bacillus subtilis NX-2. Biotechnol. Bioprocess Eng. 2011, 16, 1144–1151. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, H.; Shi, N.; Yao, J.; Li, S.; Ouyang, P. Improvement of poly(γ-glutamic acid) biosynthesis and redistribution of metabolic flux with the presence of different additives in Bacillus subtilis CGMCC 0833. Appl. Microbiol. Biotechnol. 2008, 79, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shi, F.; Zhang, B.; Zhu, F.; Cao, W.; Xu, Z.; Xu, G.; Cen, P. Effects of cultivation conditions on the production of γ-PGA with Bacillus subtilis ZJU-7. Appl. Biochem. Biotechnol. 2010, 160, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Shi, F.; Xu, Z.; Cen, P. Efficient production of poly-γ-glutamic acid by Bacillus subtilis ZJU-7. Appl. Biochem. Biotechnol. 2006, 133, 271–281. [Google Scholar] [CrossRef]
- Ju, W.T.; Song, Y.S.; Jung, W.J.; Park, R.D. Enhanced production of poly-γ-glutamic acid by a newly-isolated Bacillus subtilis. Biotechnol. Lett. 2014, 36, 2319–2324. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, I.B.; Lele, S.S.; Singhal, R.S. A statistical approach to optimization of fermentative production of poly(γ-glutamic acid) from Bacillus licheniformis NCIM 2324. Bioresour. Technol. 2009, 100, 826–832. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.B.; Cantarelli, V.V.; Ayub, M.A. Production and optimization of poly-γ-glutamic acid by Bacillus subtilis BL53 isolated from the amazonian environment. Bioprocess Biosyst. Eng. 2014, 37, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, I.B.; Singhal, R.S. Effect of aeration and agitation on synthesis of poly(γ-glutamic acid) in batch cultures of Bacillus licheniformis NCIM 2324. Biotechnol. Bioprocess Eng. 2010, 15, 635–640. [Google Scholar] [CrossRef]
- Mitsunaga, H.; Meissner, L.; Buchs, J.; Fukusaki, E. Branched chain amino acids maintain the molecular weight of poly(γ-glutamic acid) of Bacillus licheniformis ATCC 9945 during the fermentation. J. Biosci. Bioeng. 2016, 122, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.B.; Mohamed, S.A.O. Enhanced production of poly glutamic acid by Bacillus sp. SW1-2 using statistical experimental design. Afr. J. Biotechnol. 2013, 12, 481–490. [Google Scholar] [CrossRef]
- Zhang, D.; Feng, X.; Zhou, Z.; Zhang, Y.; Xu, H. Economical production of poly(γ-glutamic acid) using untreated cane molasses and monosodium glutamate waste liquor by Bacillus subtilis NX-2. Bioresour. Technol. 2012, 114, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.R.; Lee, S.M.; Cho, K.S.; Jeong, S.Y.; Hwang, D.Y.; Kim, D.S.; Hong, C.O.; Son, H.J. Improved production of poly-γ-glutamic acid by Bacillus subtilis D7 isolated from Doenjang, a korean traditional fermented food, and its antioxidant activity. Appl. Biochem. Biotechnol. 2014, 173, 918–932. [Google Scholar] [CrossRef] [PubMed]
- Zeng, W.; Li, W.; Shu, L.; Yi, J.; Chen, G.; Liang, Z. Non-sterilized fermentative co-production of poly(γ-glutamic acid) and fibrinolytic enzyme by a thermophilic Bacillus subtilis GXA-28. Bioresour. Technol. 2013, 142, 697–700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.; Zhu, X.; Cai, J.; Zhang, A.; Hong, Y.; Huang, J.; Huang, L.; Xu, Z. High-level exogenous glutamic acid-independent production of poly-(γ-glutamic acid) with organic acid addition in a new isolated Bacillus subtilis C10. Bioresour. Technol. 2012, 116, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Jiang, B.; Zhang, T.; Mu, W.; Miao, M.; Hua, Y. High-level production of poly(γ-glutamic acid) by a newly isolated glutamate-independent strain, Bacillus methylotrophicus. Process Biochem. 2015, 50, 329–335. [Google Scholar] [CrossRef]
- Tang, B.; Xu, H.; Xu, Z.; Xu, C.; Xu, Z.; Lei, P.; Qiu, Y.; Liang, J.; Feng, X. Conversion of agroindustrial residues for high poly(γ-glutamic acid) production by Bacillus subtilis NX-2 via solid-state fermentation. Bioresour. Technol. 2015, 181, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.R.; Irorere, V.U.; Bartlett, T.; Hill, D.; Kedia, G.; Morris, M.R.; Charalampopoulos, D.; Radecka, I. Bacillus subtilis natto: A non-toxic source of poly-γ-glutamic acid that could be used as a cryoprotectant for probiotic bacteria. AMB Express 2013, 3, 36. [Google Scholar] [CrossRef] [PubMed]
- Mitsuiki, M.; Mizuno, A.; Tanimoto, H.; Motoki, M. Relationship between the antifreeze activities and the chemical structures of oligo-and poly(glutamic acid)s. J. Agric. Food. Chem. 1998, 46, 891–895. [Google Scholar] [CrossRef]
- Kubota, H.; Nambu, Y.; Endo, T. Convenient esterification of poly(γ-glutamic acid) produced by microorganism with alkyl halides and their thermal properties. J. Polym. Sci. Part A Polym. Chem. 1995, 33, 85–88. [Google Scholar] [CrossRef]
- Inbaraj, B.S.; Chiu, C.; Ho, G.; Yang, J.; Chen, B. Removal of cationic dyes from aqueous solution using an anionic poly-γ-glutamic acid-based adsorbent. J. Hazard. Mater. 2006, 137, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Uotani, K.; Kubota, H.; Endou, H.; Tokita, F. Sialogogue, Oral Composition and Food Product Containing the Same. U.S. Patent 7,910,089, 22 March 2011. [Google Scholar]
- Ye, H.; Jin, L.; Hu, R.; Yi, Z.; Li, J.; Wu, Y.; Xi, X.; Wu, Z. Poly(γ, l-glutamic acid)–cisplatin conjugate effectively inhibits human breast tumor xenografted in nude mice. Biomaterials 2006, 27, 5958–5965. [Google Scholar] [CrossRef] [PubMed]
- Ogata, M.; Hidari, K.I.; Murata, T.; Shimada, S.; Kozaki, W.; Park, E.Y.; Suzuki, T.; Usui, T. Chemoenzymatic synthesis of sialoglycopolypeptides as glycomimetics to block infection by avian and human influenza viruses. Bioconjug. Chem. 2009, 20, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, I.B.; Singhal, R.S. Flocculation properties of poly(γ-glutamic acid) produced from Bacillus subtilis isolate. Food Bioprocess Technol. 2011, 4, 745–752. [Google Scholar] [CrossRef]
- Ho, G.H.; Yang, T.H.; Yang, J. Dietary Products Comprising one or more of γ-Polyglutamic Acid (γ-PGA, H Form) and γ-Polyglutamates for Use as Nutrition Supplements. U.S. Patent 7,632,804, 15 December 2009. [Google Scholar]
- Ben-zur, N.; Goldman, D.M. γ-poly glutamic acid: A novel peptide for skin care. Cosmet. Toilet. 2007, 122, 65–74. [Google Scholar]
- Hajdu, I.; Bodnár, M.; Csikós, Z.; Wei, S.; Daróczi, L.; Kovács, B.; Győri, Z.; Tamás, J.; Borbély, J. Combined nano-membrane technology for removal of lead ions. J. Membr. Sci. 2012, 409, 44–53. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, S.; Zhang, J.; Sun, M.; Liu, Z.; Yu, Z. Co-producing lipopeptides and poly-γ-glutamic acid by solid-state fermentation of Bacillus subtilis using soybean and sweet potato residues and its biocontrol and fertilizer synergistic effects. Bioresour. Technol. 2008, 99, 3318–3323. [Google Scholar] [CrossRef] [PubMed]
- Cromwick, A.M.; Birrer, G.A.; Gross, R.A. Effects of ph and aeration on γ-poly(glutamic acid) formation by Bacillus licheniformis in controlled batch fermentor cultures. Biotechnol. Bioeng. 1996, 50, 222–227. [Google Scholar] [CrossRef]





| γ-PGA | Composition (%) | Strains | Reference |
|---|---|---|---|
| d-Glutamate | 100 | Bacillus anthracis | [11] |
| l-Glutamate | 100 | Natrialba aegyptiaca | [24] |
| d-/l-Glutamate | 60/40 | Bacillus subtilis | [28,29] |
| 10–100/10–90 | Bacillus licheniformis | [25,26] | |
| 30/70 | Bacillus megaterium | [27] | |
| 40/60 | Staphylococcus epidermidis | [23] |
| Strains | Genotype | Fermentation Medium | Wild-Type Yield (g/L) | Yield (g/L) | Increasing Yield (%) | Reference |
|---|---|---|---|---|---|---|
| B. amyloliquefaciens LL3 | ΔpgdSΔcwlO | Sucrose, (NH4)2SO4, MgSO4, KH2PO4, K2HPO4 | 3.69 | 7.12 | 92.95 | [44] |
| B. amyloliquefaciens | ΔcwlOΔepsA-Ovgb | Sucrose, (NH4)2SO4, MgSO4, KH2PO4, K2HPO4, trace elements (FeSO4·4H2O, CaCl2·2H2O, MnSO4·4H2O, ZnCl2) | 3.14 | 5.12 | 63.06 | [45] |
| B. amyloliquefaciens LL3 | ΔrocRΔrocGΔgudBΔodhA | Sucrose, (NH4)2SO4, MgSO4, KH2PO4, K2HPO4 | 4.03 | 5.68 | 40.94 | [46] |
| B. subtilis PB5249 | ΔpgdSΔggt | l-glutamate, citric acid, glucose, NH4Cl, K2HPO4, MgSO4·7H2O, FeCl3·6H2O, CaCl2·2H2O, MnSO4·H2O | 20 | 40 | 100 | [47] |
| B. subtilis WB600 | pWB980-pgsBCA | Glucose, sodium glutamate, (NH4)2SO4, K2HPO4, MgSO4 | 0.134 | 1.74 | 1198.51 | [48] |
| B. subtilis subsp. chungkookjang | pWPSE-PxylA-pgsE | l-glutamate, sodium citrate, casamino acid, yeast extract, (NH4)2SO4, MgCl2, Na2HPO4, KH2PO4, NaCl | 0.20 | 0.64 | 220 | [49] |
| B. licheniformis WX-02 | pHY300PLK-P43-glr | Sucrose, (NH4)2SO4, MgSO4, KH2PO4, K2HPO4 | 11.73 | 14.38 | 22.59 | [50] |
| B. licheniformis WX-02 | pHY300PLK-PpgdS-pgdS | Glucose, sodium glutamate, sodium citrate, NH4Cl, MgSO4, K2HPO4, CaCl2, ZnSO4, MnSO4 | 13.11 | 20.16 | 53.78 | [51] |
| B. subtilis ISW1214 | pWH1520-PxylA-pgsBCA | Sucrose, NaCl, MgSO4, KH2PO4, NaHPO4, xylose | 8.2 | 9.0 | 9.76 | [52] |
| B. amyloliquefaciens | sRNA of rocG (repressed rocG and glnA genes) | Sucrose, (NH4)2SO4, MgSO4, KH2PO4, K2HPO4 | 14.96 | 20.3 | 35.69 | [53] |
| Main Substrate | Strain | Recipe | Fermentation Conditions | Flask/Fermenter | Yield (g/L) | Reference |
|---|---|---|---|---|---|---|
| Glucose + Glutamate | B. subtilis NX-2 | Glucose, l-glutamate, MgSO4, K2HPO4·3H2O, NH4SO4, MnSO2 | 7.5-L bioreactor, 400 rpm, 1.2 vvm, pH 7.0, 32 °C | Fermenter | 71.21 | [69] |
| B. subtilis NX-2 | Glucose, l-glutamate, Glycerol, K2HPO4·3H2O, MgSO4, (NH4)2SO4, MnSO4 | 500-mL flask, 220 rpm, pH 7.5, 32.5 °C | Flask | 31.7 | [62] | |
| B. subtilis NX-2 | Glucose, l-glutamate, K2HPO4·3H2O, MgSO4, NH4Cl | 110-L bioreactor, 220 rpm, pH 7.5, 32.5 °C | Fermenter | 35.0 | [70] | |
| B. subtilis NX-2 | Glucose, l-glutamate, (NH4)2SO4, K2HPO4·3H2O, MgSO4, MnSO4 | 7.5-L BioFlo 110 bioreactor, 200–800 rpm, 1.5 vvm, pH 7.0, 32 °C | Fermenter | 40.5 | [71] | |
| B. subtilis ZJU-7 | Glucose, l-glutamate, yeast extract, NaCl, CaCl2, MgSO4, MnSO4 | 10-L bioreactor, 300–800 rpm, 1.5 vvm, pH 6.5, 37 °C | Fermenter | 101.1 | [60] | |
| B. subtilis ZJU-7 | Glucose, l-glutamate, tryptone, NaCl, MgSO4, CaCl2 | 500-mL flask, 200 rpm, pH 7.0, 37 °C | Flask | 58.2 | [65] | |
| B. subtilis CGMCC 0833 | Glucose, l-glutamate, (NH4)2SO4, K2HPO4·3H2O, MgSO4, MnSO4 | 7.5-L reactor, 400 rpm, pH 7.5, 32.5 °C | Fermenter | 34.4 | [72] | |
| B. subtilis ZJU-7 | Glucose, l-glutamate, (NH4)2SO4, K2HPO4·3H2O, MnSO4, MgSO4 | 100-L fermenter, 200–450 rpm, 0.5–1 vvm, pH 6.5, 30 °C | Fermenter | 54.0 | [73] | |
| B. licheniformis P-104 | Glucose, sodium glutamate, sodium citrate, (NH4)2SO4, MnSO4, MgSO4, K2HPO4, NaNO3 | 7-L bioreactor, 500 rpm, 1.5 vvm, pH 7.2 | Fermenter | 41.6 | [14] | |
| B. subtilis ZJU-7 | Glucose, l-glutamate, tryptone, NaCl | 7.0, 37 °C, fed-batch 100 mL flask, 200 rpm, pH 7.0, 37 °C | Flask | 54.4 | [74] | |
| Citrate + Glutamate | B. licheniformis ATCC9945 | Citric acid, l-glutamate, glycerol, NH4Cl, K2HPO4, MgSO4·7H2O, FeCl3·6H2O, CaCl2·2H2O, MnSO4·H2O | 225 mL flask, 250 rpm, pH 6.5, 30 °C | Flask | 12.64 | [64] |
| B. subtilis MJ80 | Citric acids, l-glutamate, starch, urea, glycerol | 300 L fermenter, 150 rpm, 1 vvm, initial pH 7.0, 37 °C | Fermenter | 68.7 | [75] | |
| B. licheniformis NCIM 2324 | Citric acid, l-glutamate, glycerol, (NH4)2SO4, K2HPO4, MgSO4·7H2O, MnSO4·7H2O, CaCl2·2H2O, MnSO4·7H2O | 250-mL flask, 200 rpm, initial pH 6.5, 37 °C | Flask | 35.75 | [76] | |
| B. licheniformis NCIM 2324 | Citric acids, l-glutamate, MgSO4·7H2O, MnSO4·2H2O α-ketoglutaric acid | 250-mL Erlenmeyer flask, 200 rpm, pH 7.0, 37 °C | Flask | 98.64 | [63] | |
| B. subtilis BL53 | Citric acid, l-glutamate, glycerol, NH4Cl, MgSO4·7H2O, FeCl3·6H2O, K2HPO4, CaCl2·2H2O, MnSO4·H2O | 250-mL flask, 180 rpm, pH 6.5, 37 °C | Flask | 17.0 | [77] | |
| B. licheniformis NCIM 2324 | Citric acids, l-glutamate glycerol, ammonium sulphate | 250-mL flasks, 200 rpm, pH 5–8, 37 °C | Flask | 26.12 | [17] | |
| B. licheniformis NCIM 2324 | Citric acid, l-glutamate, glycerol, ammonium sulfate, α-ketoglutaric acid, K2HPO4, MgSO4·7H2O, CaCl2·2H2O, MnSO4·7H2O | Biostat B5 fermenter (2.5 L), 250–1000 rpm, 1.0–3.0 vvm, pH 6.5, 37 °C | Fermenter | 46.34 | [78] | |
| B. licheniformis ATCC 9945 | Citric acid, l-glutamate, glycerol, NH4Cl, K2HPO4, MgSO4·7H2O, FeCl3·6H2O, CaCl2·2H2O, MnSO4·H2O | 500-mL Erlenmeyer flask, pH 7.4, 37 °C | Flask | 35.78 | [79] | |
| B. sp. SW1-2 | Citric acids, l-glutamate, glycerol, K2HPO4, MgSO4·7H2O, FeSO4·4H2O, CaCl2·2H2O, MnSO4·4H2O, ZnCl2, NH4)2SO4, casein hydrolysate. | 250-mL Erlenmeyer flask, 150 rpm, pH 7.0, 37 °C | Flask | 36.5 | [80] | |
| Other sugars + Glutamate | B. subtilis NX-2 | l-glutamate, (NH4)2SO4, K2HPO4, MgSO4, MnSO4, carbon source (glucose, xylose) | 7.5-L bioreactor, 400 rpm, 1.2 vvm, initial pH 7.0, 32 °C | Fermenter | 73.0 | [61] |
| B. subtilis HB-1 | l-glutamate, xylose, yeast extract, NaCl, MgSO4, CaCl2, | 10-L bioreactor, 500 rpm, initial pH 6.5, 37 °C, fed-batch | Fermenter | 28.15 | [66] | |
| B. subtilis NX-2 | Cane molasses, glutamate, (NH4)2SO4, wet cells, crude protein, reducing sugar, potassium, calcium, magnesium, manganese, iron, phosphonium. | 7.5-L bioreactor, 400 rpm, 1.2 vvm, pH 7.0, 32 °C | Fermenter | 52.1 | [81] | |
| B. subtilis D7 | l-glutamate acid, mannitol, yeast extract, K2HPO4, NaH2PO4, MgSO4·7H2O, CuSO4·5H2O, pyruvic acid | 300-mL Erlenmeyer flask, 250 rpm, pH 8.0, 35 °C | Flask | 24.93 | [82] | |
| B. subtilis GXA-28 | soybean residue, sucrose, glutamate | 250-mL flask, 200 rpm, pH 2.0–10.0, 45 °C | Flask | 8.72 | [83] | |
| Glucose + NH4Cl | B. subtilis C10 | Glucose, NH4Cl, MgSO4·7H2O, K2HPO4, FeCl3·6H2O, MgSO4·H2O, CaCl2, CaCO3 | 10-L fermenter, 200–500 rpm, 1.5 vvm, pH 7.5, 32 °C | Fermenter | 27.70 | [84] |
| B. licheniformis A13 | Glucose, NH4Cl, NaCl, MgSO4·7H2O, CaCl2·2H2O, K2HPO4, FeSO4·4H2O, NaMoO4, CuSO4, MnSO4, ZnSO4, CoCl2, H3BO4 | 250-mL Erlenmeyer flask, 200 rpm, pH 6.5, 37 °C | Flask | 28.2 | [67] | |
| B. licheniformis TISTR 1010 | Glucose, citric acid, NH4Cl, K2HPO4, MgSO4·7H2O, FeCl3·6H2O, CaCl2·2H2O, MnSO4·H2O, NaCl, Tween-80 | 7-L fermenter, 300 rpm, 1 vvm, pH 7.4, 37 °C | Fermenter | 27.50 | [68] | |
| Others | B. methylotrophicus | Glucose, yeast extract, MgSO4·7H2O, K2HPO4 MnSO4 | 250-mL flasks, 200 rpm, pH 7.2, 37 °C | Flask | 35.34 | [85] |
| B. subtilis NX-2 | Glucose, cane molasses, xylose, starch, industrial waste glycerol, citric acid, DMR, MGPR (oyster, shiitake, needle, eryngii mushroom, and Agaricus bisporus residues | 500-mL shake flask, 150 rpm, pH 7.0, 35 °C | Flask | 107.7 | [86] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsueh, Y.-H.; Huang, K.-Y.; Kunene, S.C.; Lee, T.-Y. Poly-γ-glutamic Acid Synthesis, Gene Regulation, Phylogenetic Relationships, and Role in Fermentation. Int. J. Mol. Sci. 2017, 18, 2644. https://doi.org/10.3390/ijms18122644
Hsueh Y-H, Huang K-Y, Kunene SC, Lee T-Y. Poly-γ-glutamic Acid Synthesis, Gene Regulation, Phylogenetic Relationships, and Role in Fermentation. International Journal of Molecular Sciences. 2017; 18(12):2644. https://doi.org/10.3390/ijms18122644
Chicago/Turabian StyleHsueh, Yi-Huang, Kai-Yao Huang, Sikhumbuzo Charles Kunene, and Tzong-Yi Lee. 2017. "Poly-γ-glutamic Acid Synthesis, Gene Regulation, Phylogenetic Relationships, and Role in Fermentation" International Journal of Molecular Sciences 18, no. 12: 2644. https://doi.org/10.3390/ijms18122644