Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury
Abstract
:1. Introduction
2. Results
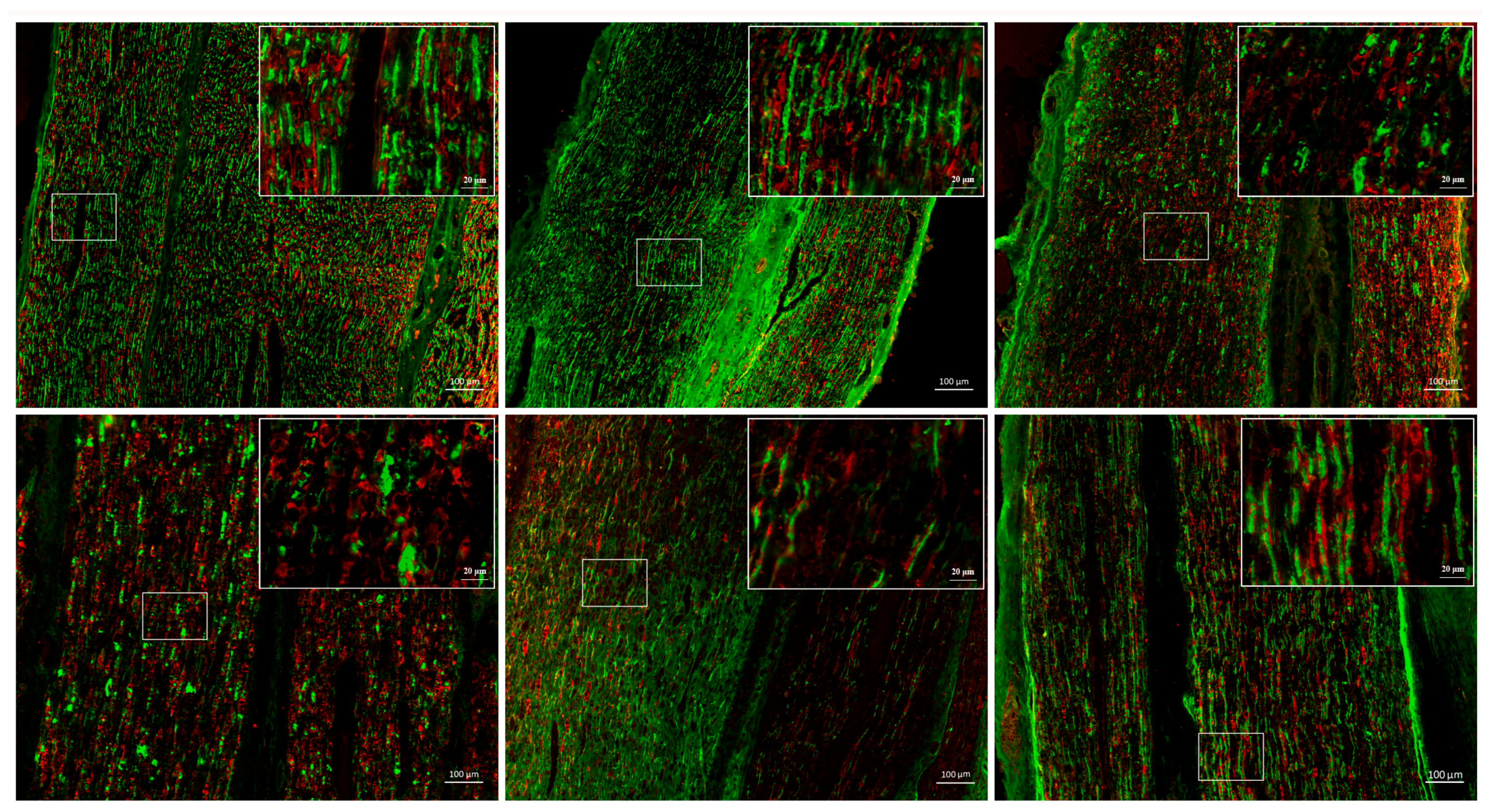
2.1. Peripheral Nerve Injury Involves Complex Biological Processes
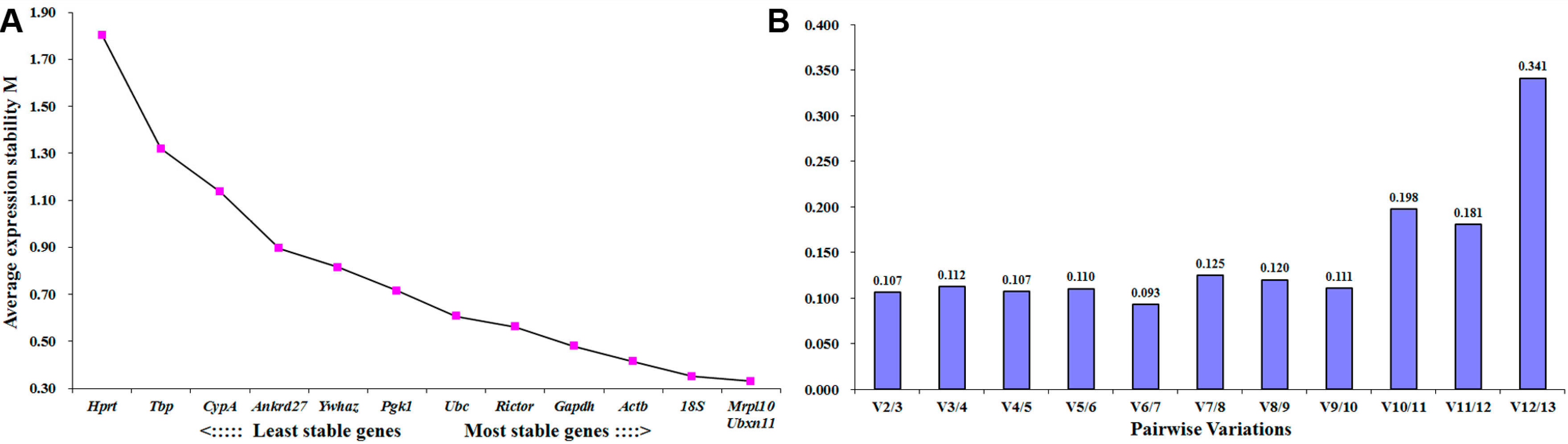
2.2. Mrpl10 Is the Most Stable Expressed Reference Gene in Distal Sciatic Nerve Following Peripheral Nerve Injury
2.3. Tbp Is the Most Stable Expressed Reference Gene in DRGs Following Peripheral Nerve Injury
3. Discussion
4. Materials and Methods
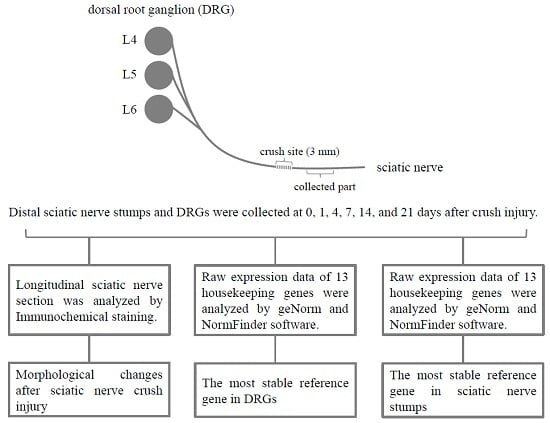
4.1. Animal Surgery
4.2. Immunochemical Staining
4.3. RNA Extraction
4.4. qRT-PCR
4.5. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bosse, F. Extrinsic cellular and molecular mediators of peripheral axonal regeneration. Cell Tissue Res. 2012, 349, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Fournier, A.E.; Strittmatter, S.M. Regenerating nerves follow the road more traveled. Nat. Neurosci. 2002, 5, 821–822. [Google Scholar] [CrossRef] [PubMed]
- Glenn, T.D.; Talbot, W.S. Signals regulating myelination in peripheral nerves and the Schwann cell response to injury. Curr. Opin. Neurobiol. 2013, 23, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Christie, K.J.; Zochodne, D. Peripheral axon regrowth: New molecular approaches. Neuroscience 2013, 240, 310–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.L.; Yu, W.M.; Strickland, S. Peripheral regeneration. Annu. Rev. Neurosci. 2007, 30, 209–233. [Google Scholar] [CrossRef] [PubMed]
- Saiki, R.K.; Scharf, S.; Faloona, F.; Mullis, K.B.; Horn, G.T.; Erlich, H.A.; Arnheim, N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 1985, 230, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Nolan, T.; Pfaffl, M.W. Quantitative real-time RT-PCR—A perspective. J. Mol. Endocrinol. 2005, 34, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-Time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Butte, A.J.; Dzau, V.J.; Glueck, S.B. Further defining housekeeping, or “maintenance,” genes Focus on “A compendium of gene expression in normal human tissues”. Physiol. Genom. 2001, 7, 95–96. [Google Scholar]
- Zhu, J.; He, F.; Hu, S.; Yu, J. On the nature of human housekeeping genes. Trends Genet. 2008, 24, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Greer, S.; Honeywell, R.; Geletu, M.; Arulanandam, R.; Raptis, L. Housekeeping genes; expression levels may change with density of cultured cells. J. Immunol. Methods 2010, 355, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Gambarotta, G.; Ronchi, G.; Friard, O.; Galletta, P.; Perroteau, I.; Geuna, S. Identification and validation of suitable housekeeping genes for normalizing quantitative real-time PCR assays in injured peripheral nerves. PLoS ONE 2014, 9, e105601. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; de Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; de Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, K.; Smeets, K.; Mulder, M.; Hendriks, J.J.; Ameloot, M. Selection of reference genes for gene expression studies in rat oligodendrocytes using quantitative real time PCR. J. Neurosci. Methods 2010, 187, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Dheda, K.; Huggett, J.F.; Bustin, S.A.; Johnson, M.A.; Rook, G.; Zumla, A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 2004, 37, 112–114, 116, 118–119. [Google Scholar] [PubMed]
- Yu, J.; Gu, X.; Yi, S. Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: Insights into Wallerian degeneration. Front. Cell. Neurosci. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.; Zhang, H.; Gong, L.; Wu, J.; Zha, G.; Zhou, S.; Gu, X.; Yu, B. Deep Sequencing and Bioinformatic Analysis of Lesioned Sciatic Nerves after Crush Injury. PLoS ONE 2015, 10, e0143491. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Li, M.; Shen, D.; Ding, F.; Lu, S.; Zhao, Q.; Gu, X. Gene expression profiling of the rat sciatic nerve in early Wallerian degeneration after injury. Neural Regen. Res. 2012, 7, 1285–1292. [Google Scholar] [PubMed]
- Yao, D.; Li, M.; Shen, D.; Ding, F.; Lu, S.; Zhao, Q.; Gu, X. Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci. Bull. 2013, 29, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Wu, J.; Zhou, S.; Wang, Y.; Qin, J.; Yu, B.; Gu, X.; Yao, C. Global analysis of transcriptome in dorsal root ganglia following peripheral nerve injury in rats. Biochem. Biophys. Res. Commun. 2016, 478, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xue, C.; Yuan, Y.; Zhang, R.; Wang, Y.; Yu, B.; Liu, J.; Ding, F.; Yang, Y.; Gu, X. The transcriptional landscape of dorsal root ganglia after sciatic nerve transection. Sci. Rep. 2015, 5, 16888. [Google Scholar] [CrossRef] [PubMed]
- Bangaru, M.L.; Park, F.; Hudmon, A.; McCallum, J.B.; Hogan, Q.H. Quantification of gene expression after painful nerve injury: Validation of optimal reference genes. J. Mol. Neurosci. 2012, 46, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Beamonte, R.; Navarro, M.A.; Larraga, A.; Strunk, M.; Barranquero, C.; Acín, S.; Guzman, M.A.; Iñigo, P.; Osada, J. Selection of reference genes for gene expression studies in rats. J. Biotechnol. 2011, 151, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Seol, D.; Choe, H.; Zheng, H.; Jang, K.; Ramakrishnan, P.S.; Lim, T.H.; Martin, J.A. Selection of reference genes for normalization of quantitative real-time PCR in organ culture of the rat and rabbit intervertebral disc. BMC Res. Notes 2011, 4, 162. [Google Scholar] [CrossRef] [PubMed]
- Langnaese, K.; John, R.; Schweizer, H.; Ebmeyer, U.; Keilhoff, G. Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. BMC Mol. Biol. 2008, 9, 53. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | 0 Day | 1 Day | 4 Days | 7 Days | 14 Days | 21 Days |
|---|---|---|---|---|---|---|
| 18S | 9.27 ± 0.77 | 8.18 ± 0.28 | 5.33 ± 0.26 | 5.43 ± 0.34 | 5.51 ± 0.30 | 8.67 ± 0.29 |
| Actb | 19.82 ± 0.28 | 17.90 ± 0.09 | 16.02 ± 0.10 | 16.44 ± 0.16 | 16.89 ± 0.08 | 19.37 ± 0.39 |
| Ankrd27 | 28.97 ± 0.74 | 29.13 ± 0.36 | 24.36 ± 0.19 | 24.37 ± 0.18 | 24.15 ± 0.18 | 28.30 ± 0.59 |
| CypA | 25.46 ± 0.70 | 24.21 ± 0.54 | 17.87 ± 0.32 | 18.26 ± 0.33 | 19.14 ± 0.55 | 26.19 ± 0.70 |
| Gapdh | 21.05 ± 0.46 | 19.95 ± 0.02 | 17.73 ± 0.12 | 18.77 ± 0.08 | 19.04 ± 0.12 | 21.68 ± 0.08 |
| Hprt | 35.81 ± 0.22 | 30.93 ± 0.04 | 21.74 ± 0.74 | 21.77 ± 0.85 | 22.01 ± 0.69 | 33.10 ± 0.84 |
| Mrpl10 | 28.95 ± 0.64 | 27.90 ± 0.09 | 25.41 ± 0.24 | 25.86 ± 0.22 | 26.05 ± 0.12 | 28.81 ± 0.12 |
| Pgk1 | 25.62 ± 0.59 | 23.26 ± 0.45 | 20.19 ± 0.47 | 21.18 ± 0.51 | 22.12 ± 0.49 | 25.61 ± 0.47 |
| Rictor | 26.93 ± 0.00 | 26.64 ± 0.29 | 24.92 ± 0.16 | 24.87 ± 0.19 | 24.84 ± 0.17 | 26.53 ± 0.22 |
| Tbp | 32.71 ± 0.12 | 30.54 ± 0.54 | 24.77 ± 0.43 | 24.75 ± 0.44 | 24.69 ± 0.44 | 32.57 ± 0.61 |
| Ubc | 22.94 ± 0.00 | 22.84 ± 0.21 | 20.79 ± 0.22 | 21.13 ± 0.21 | 21.29 ± 0.28 | 22.43 ± 0.24 |
| Ubxn11 | 31.07 ± 0.56 | 30.54 ± 0.20 | 28.21 ± 0.22 | 28.03 ± 0.16 | 28.03 ± 0.12 | 30.94 ± 0.01 |
| Ywhaz | 24.61 ± 0.85 | 24.06 ± 0.21 | 20.21 ± 0.13 | 20.07 ± 0.01 | 20.29 ± 0.13 | 25.34 ± 0.45 |
| Ranking Order | Gene Name | Stability Value |
|---|---|---|
| 1 | Mrpl10 | 0.053 |
| 2 | 18S | 0.058 |
| 3 | Rictor | 0.097 |
| 4 | Ubc | 0.105 |
| 5 | Actb | 0.111 |
| 6 | Ywhaz | 0.112 |
| 7 | Hprt | 0.120 |
| 8 | CypA | 0.121 |
| 9 | Ubxn11 | 0.125 |
| 10 | Ankrd27 | 0.145 |
| 11 | Tbp | 0.160 |
| 12 | Gapdh | 0.202 |
| 13 | Pgk1 | 0.214 |
| Gene Name | 0 Day | 1 Day | 4 Days | 7 Days | 14 Days | 21 Days |
|---|---|---|---|---|---|---|
| 18S | 6.17 ± 0.23 | 5.72 ± 0.15 | 5.75 ± 0.14 | 5.78 ± 0.17 | 5.88 ± 0.09 | 5.90 ± 0.17 |
| Actb | 18.52 ± 0.03 | 17.94 ± 0.03 | 17.87 ± 0.10 | 17.70 ± 0.06 | 17.83 ± 0.05 | 17.87 ± 0.06 |
| Ankrd27 | 24.20 ± 0.15 | 24.56 ± 0.04 | 24.06 ± 0.17 | 23.84 ± 0.15 | 23.87 ± 0.11 | 23.93 ± 0.03 |
| CypA | 17.81 ± 0.17 | 17.57 ± 0.06 | 17.67 ± 0.12 | 17.84 ± 0.13 | 17.89 ± 0.10 | 17.91 ± 0.05 |
| Gapdh | 17.38 ± 0.76 | 17.34 ± 0.67 | 17.16 ± 0.56 | 17.13 ± 0.60 | 17.22 ± 0.61 | 16.99 ± 0.76 |
| Hprt | 23.42 ± 0.12 | 22.96 ± 0.01 | 22.48 ± 0.16 | 22.27 ± 0.08 | 22.42 ± 0.08 | 22.77 ± 0.13 |
| Mrpl10 | 25.90 ± 0.04 | 25.54 ± 0.07 | 25.49 ± 0.08 | 25.63 ± 0.17 | 25.95 ± 0.02 | 25.90 ± 0.06 |
| Pgk1 | 21.10 ± 0.07 | 20.86 ± 0.10 | 20.64 ± 0.05 | 20.35 ± 0.12 | 20.77 ± 0.09 | 20.93 ± 0.00 |
| Rictor | 25.25 ± 0.54 | 25.32 ± 0.57 | 25.14 ± 0.62 | 25.23 ± 0.62 | 25.17 ± 0.55 | 25.29 ± 0.49 |
| Tbp | 26.06 ± 0.13 | 25.90 ± 0.06 | 25.89 ± 0.04 | 25.83 ± 0.20 | 25.85 ± 0.14 | 25.92 ± 0.02 |
| Ubc | 21.86 ± 0.12 | 21.52 ± 0.08 | 21.61 ± 0.13 | 21.73 ± 0.07 | 21.91 ± 0.03 | 21.91 ± 0.03 |
| Ubxn11 | 30.23 ± 0.33 | 30.21 ± 0.29 | 29.60 ± 0.39 | 29.65 ± 0.21 | 29.74 ± 0.16 | 30.18 ± 0.43 |
| Ywhaz | 18.94 ± 0.02 | 18.92 ± 0.09 | 18.90 ± 0.06 | 18.91 ± 0.06 | 18.94 ± 0.03 | 18.88 ± 0.03 |
| Ranking Order | Gene Name | Stability Value |
|---|---|---|
| 1 | Tbp | 0.026 |
| 2 | 18S | 0.058 |
| 3 | Rictor | 0.066 |
| 4 | Ywhaz | 0.070 |
| 5 | Pgk1 | 0.083 |
| 6 | Gapdh | 0.083 |
| 7 | Actb | 0.089 |
| 8 | Mrpl10 | 0.103 |
| 9 | Ubc | 0.107 |
| 10 | CypA | 0.114 |
| 11 | Ubxn11 | 0.116 |
| 12 | Ankrd27 | 0.140 |
| 13 | Hprt | 0.144 |
| Symbol | Accession Number | Primer Sequences | Amplicon Size | Efficiency | Reference |
|---|---|---|---|---|---|
| 18S | X01117.1 | Sense: ACTCAACACGGGAAACCTCA Antisense: AATCGCTCCACCAACTAAGA | 114 | 96% | [26] |
| Actb | NM_031144.2 | Sense: AGGCCAACCGTGAAAAGATG Antisense: ACCAGAGGCATACAGGGACAA | 101 | 94% | [27] |
| Ankrd27 | NM_001271264.1 | Sense: CCAGGAGACAGAACACGAGG Antisense: CCCCTGGGTTAATGAGGCAA | 119 | 91% | * |
| CypA | NM_017101.1 | Sense: CCAAACACAAATGGTTCCCAGT Antisense: ATTCCTGGACCCAAAACGCT | 135 | 101% | [26] |
| Gapdh | NM_017008.4 | Sense: CAACTCCCTCAAGATTGTCAGCAA Antisense: GGCATGGACTGTGGTCATGA | 118 | 92% | [28] |
| Hprt | NM_012583.2 | Sense: TCCCAGCGTCGTGATTAGTGA Antisense: CCTTCATGACATCTCGAGCAAG | 152 | 91% | [26] |
| Mrpl10 | NM_001109620.1 | Sense: CTCCTCCCAAGCCCCCCAAG Antisense: CAGACAGCTATCATTCGGTTGTCCC | 97 | 92% | [13] |
| Pgk1 | NM_053291.3 | Sense: ATGCAAAGACTGGCCAAGCTAC Antisense: AGCCACAGCCTCAGCATATTTC | 104 | 91% | [28] |
| Rictor | XM_001055633.3 | Sense: GAGGTGGAGAGGACACAAGCCC Antisense: GGCCACAGAACTCGGAAACAAGG | 81 | 91% | [13] |
| Tbp | NM_001004198.1 | Sense: TGGGATTGTACCACAGCTCCA Antisense: CTCATGATGACTGCAGCAAACC | 131 | 95% | [28] |
| Ubc | NM_017314.1 | Sense: ATCTAGAAAGAGCCCTTCTTGTGC Antisense: ACACCTCCCCATCAAACCC | 51 | 90% | [26] |
| Ubxn11 | NM_138853.2 | Sense: GCGAGACTGGATGAAGGCCAAG Antisense: CCCTCCACCACCAGCTCACTC | 120 | 110% | [13] |
| Ywhaz | NM_013011.3 | Sense: GATGAAGCCATTGCTGAACTTG Antisense: GTCTCCTTGGGTATCCGATGTC | 117 | 94% | [28] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Shan, Q.; Meng, Y.; Pan, J.; Yi, S. Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury. Int. J. Mol. Sci. 2017, 18, 263. https://doi.org/10.3390/ijms18020263
Wang Y, Shan Q, Meng Y, Pan J, Yi S. Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury. International Journal of Molecular Sciences. 2017; 18(2):263. https://doi.org/10.3390/ijms18020263
Chicago/Turabian StyleWang, Yaxian, Qianqian Shan, Yali Meng, Jiacheng Pan, and Sheng Yi. 2017. "Mrpl10 and Tbp Are Suitable Reference Genes for Peripheral Nerve Crush Injury" International Journal of Molecular Sciences 18, no. 2: 263. https://doi.org/10.3390/ijms18020263