Lannea coromandelica (Houtt.) Merr. Induces Heme Oxygenase 1 (HO-1) Expression and Reduces Oxidative Stress via the p38/c-Jun N-Terminal Kinase–Nuclear Factor Erythroid 2-Related Factor 2 (p38/JNK–NRF2)-Mediated Antioxidant Pathway
Abstract
:1. Introduction
2. Results
2.1. High-Pressure Liquid Chromatography (HPLC) Analysis of Lannea coromandelica Bark Extract
2.2. Radical-Scavenging Activities of LCB Extract
2.3. LCB Extract Attenuates Cellular Oxidative Stress Induced by 2,2′-Azobis(2-Amidinopropane) Dihydrochloride (AAPH) in RAW 264.7 Cells
2.4. Effects of LCB Extract on Antioxidant Enzyme Expression
2.5. Effects of LCB Extract on Phase II Enzymes through NRF2 Nuclear Translocation
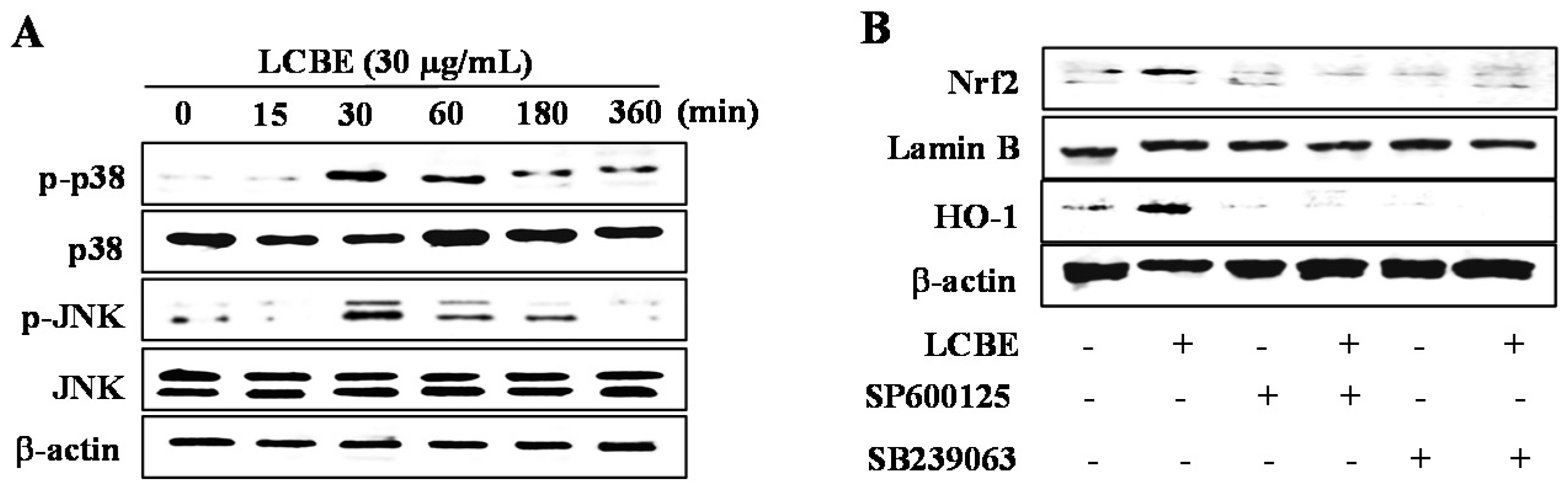
2.6. LCB Extract Activates NRF2 via Phosphorylation of JNK and p38 MAPKs
3. Discussion
4. Experimental Section
4.1. Plant Material and Extraction
4.2. Drugs and Chemicals
4.3. High-Pressure Liquid Chromatography with Diode-Array Detection Analysis
4.4. Radical-Scavenging Activity Assays
4.4.1. 2,2-Diphenyl-1-Picrylhydrazyl Radical-Scavenging Assay
4.4.2. 2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulphonic acid) Radical-Scavenging Assay
4.4.3. Superoxide Radical-Scavenging Assay
4.4.4. Hydroxyl Radical-Scavenging Assay
4.4.5. Ferric Reducing Antioxidant Power Assay
4.4.6. Cupric Reducing Antioxidant Capacity
4.4.7. Oxygen Radical Absorbance Capacity
4.5. In Silico Toxic Risk Prediction and Biological Activity Spectrum
4.6. Cell Culture and Cell Viability Assay
4.7. Measurement of Intracellular ROS
4.8. Reverse Transcription-Polymerase Chain Reaction
4.9. Preparation of Cell Lysates and Western Blotting
4.10. Statistical Data Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liang, L.; Gao, C.; Luo, M.; Wang, W.; Zhao, C.; Zu, Y.; Efferth, T.; Fu, Y. Dihydroquercetin (DHQ) induced HO-1 and NQO1 expression against oxidative stress through the Nrf2-dependent antioxidant pathway. J. Agric. Food Chem. 2013, 61, 2755–2761. [Google Scholar] [CrossRef] [PubMed]
- Bryan, H.K.; Olayanju, A.; Goldring, C.E.; Park, B.K. The Nrf2 cell defense pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013, 85, 705–717. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Zhang, D.D. Ectodermal-neural cortex 1 down-regulates Nrf2 at the translation level. PLoS ONE 2009, 4, c5492. [Google Scholar]
- Cullinan, S.B.; Gordan, J.D.; Jin, J. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell Biol. 2000, 24, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- Taguhi, K.; Motohashi, H.; Yamamoto, M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 2011, 16, 123–140. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Dinkova-Kostova, A.T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol. 2011, 85, 241–272. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Lemos, M.; Sharma, M.; Shriram, V. Antioxidant and DNA damage protecting activities of Eulophia nuda Lindl. Free Radic. Antiox. 2013, 3, 55–60. [Google Scholar] [CrossRef]
- Chun, K.; Alam, M.B.; Son, H.U.; Lee, S.H. Effect of novel compound LX519290, a derivative of L-allo threonine, on antioxidant potential in vitro and in vivo. Int. J. Mol. Sci. 2016, 17, 1451. [Google Scholar] [CrossRef] [PubMed]
- Rahmatullah, M.; Azam, M.N.; Khatun, Z.; Seraj, S.; Islam, F.; Rahman, M.A.; Jahan, S.; Aziz, M.S. Medicinal plants used for treatment of diabetes by the Marakh sect of the Garo tribe living in Mymensingh district, Bangladesh. Afr. J. Tradit. Complement Altern. Med. 2012, 9, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.L.; Xing, F.W. Ethnobotanical study on medicinal plants around Mt. Yinggeling, Hainan Island, China. J. Ethnopharmacol. 2009, 124, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Mulaudzi, R.B.; Ndhlala, A.R.; Kulkarni, M.G.; van Staden, J. Pharmacological properties and protein binding capacity of phenolic extracts of some Venda medicinal plants used against cough and fever. J. Ethnopharmacol. 2012, 143, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Kadir, M.F.; Sayeed, M.S.B.; Mia, M.M.K. Ethnopharmacological survey of medicinal plants used by traditional healers in Bangladesh for gastrointestinal disorders. J. Ethnopharmacol. 2013, 147, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Imam, M. Z.; Moniruzzaman, M. Antinociceptive effect of ethanol extract of leaves of Lannea coromandelica. J. Ethnopharmacol. 2014, 154, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Weerapreeyakul, N.; Junhom, C.; Barusrux, S.; Thitimetharoch, T. Induction of apoptosis in human hepatocellular carcinoma cells by extracts of Lannea coromandelica (Houtt.) Merr. and Diospyros castanea (Craib) Fletcher. Chin. Med. 2016, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Egbe, E.O.; Akumka, D.D.; Adamu, M.; Mikail, H.G. Phytochemistry, antinociceptive and anti-inflammatory actvities of methanolic leaves extract of Lannea schimperi (Hoschst. Ex Rich) ENG. Recent Pat. Biotechnol. 2016, 9, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.; Jain, V. Appraisal of total phenol, flavonoid contents, and antioxidant potential of folkloric Lannea coromandelica using in vitro and in vivo assays. Scientifica 2015, 2015, 203679. [Google Scholar] [CrossRef] [PubMed]
- Kaur, R.; Jaiswal, M.L.; Jain, V. Protective effect of Lannea coromandelica Houtt. Merrill. against three common pathogens. J. Ayurveda Integr. Med. 2013, 4, 224–228. [Google Scholar] [PubMed]
- Singh, S.; Singh, G. Anti-inflammatory activity of Lannea coromandelica bark extract in rats. Phytother. Res. 1994, 8, 311–313. [Google Scholar] [CrossRef]
- Singh, S.; Singh, G. Hypotensive activity of Lannea coromandelica bark extract. Phytother. Res. 1996, 10, 429–430. [Google Scholar] [CrossRef]
- Alam, B.; Hossain, S.; Habib, R.; Rea, J.; Islam, A. Antioxidant and analgesic activities of Lannea coromandelica Linn. bark extract. Int. J. Pharmacol. 2012, 8, 224–233. [Google Scholar]
- Islam, M.T.; Ito, T.; Sakasai, M.; Tahara, S. Zoosporicidal activity of polyflavonoid tannin identified in Lannea coromandelica stem bark against phytopathogenic oomycete Aphanomyces cochlioides. J. Agric. Food Chem. 2002, 50, 6697–6703. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-kostova, A.T.; Talalay, P. Direct and indirect antioxidant properties of inducers of cytoprotective proteins. Mol. Nutr. Food Res. 2008, 52, S128–S138. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, N.; Yamashita, H.; Gotoh, N.; Yamamoto, Y.; Numano, R.; Niki, E. 2,29-Azobis (4-Methoxy-2,4-Dimethylvaleronitrile), a new lipid-soluble azo Iinitiator: Application to oxidations of lipids and low-density lipoprotein in solution and in aqueous dispersions. Free Radic. Biol. Med. 1998, 24, 259–268. [Google Scholar] [CrossRef]
- Liu, X.P.; Goldring, C.E.P.; Copple, I.M.; Wang, H.Y.; Wei, W.; Kitteringham, N.R.; Park, B.K. Extract of Ginkgo biloba induces phase 2 genes through Keap1-Nrf2-ARE signaling pathway. Life Sci. 2007, 80, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef] [PubMed]
- Reczek, C.R.; Chandel, N.S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2014, 33C, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Kumaran, A.; Karunakaran, R.J. In vitro antioxidant activities of methanol extracts of five Phyllanthus species from India. LWT Food Sci. Technol. 2007, 40, 344–352. [Google Scholar] [CrossRef]
- Kahkonen, M.P.; Hopia, A.I.; Vuorela, H.J.; Rauha, J.P.; Pihlaja, K.; Kujala, T.S.; Heinonen, M. Antioxidant activity of plant extracts containing phenolic compounds. J. Agric. Food Chem. 2007, 47, 3954–3962. [Google Scholar] [CrossRef]
- Holasova, M.; Fiedlerova, V.; Smrcinova, H.; Orsak, M.; Lachman, J.; Vavreinova, S. Buckwheat—The source of antioxidant activity in functional foods. Food Res. Int. 2002, 35, 207–211. [Google Scholar] [CrossRef]
- Gheldof, N.; Engeseth, N.J. Antioxidant capacity of honeys from various floral sources based on the determination of oxygen radical absorbance capacity and inhibition of in vitro lipoprotein oxidation in human serum samples. J. Agric. Food Chem. 2002, 50, 3050–3055. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.S.; Nair, A.G.R. Polyphenols of Lannea coromandelica. Phytochemistry 1971, 10, 1939–1940. [Google Scholar] [CrossRef]
- Islam, M.T.; Tahara, S. Dihydroflavonols from Lannea coromandelica. Phytochemistry 2000, 54, 901–907. [Google Scholar] [CrossRef]
- Wollenweber, E.; Dietz, V.H. Occurrence and distribution of free flavonoid aglycones in plants. Phytochemistry 1981, 20, 869. [Google Scholar] [CrossRef]
- Na, H.K.; Kim, E.H.; Jung, J.H.; Lee, H.H.; Hyun, J.W.; Surh, Y.J. (−)-Epigallocatechin gallate induces Nrf2-mediated antioxidant enzyme expression via activation of PI3K and ERK in human mammary epithelial cells. Arch Biochem. Biophys. 2008, 476, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.P.; Yun, H.J.; Chun, H.K.; Chung, Y.C.; Kim, H.K.; Jeong, M.H.; Yoon, T.R.; Jeong, H.G. Protective mechanisms of 3-caffeoyl, 4-dihydrocaffeoyl quinic acid from Salicornia herbacea against tert-butyl hydroperoxide-induced oxidative damage. Chem. Biol. Interact. 2009, 181, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Ou, B. The chemistry behind antioxidant capacity assays. J. Agric. Food Chem. 2005, 53, 1841–1856. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, P.M.; Prabhakar, P.K.; Doble, M. Synthesis, antioxidant evaluation and quantitative structure-activity relationship studies of chalcones. Med. Chem. Res. 2010, 19, 1–17. [Google Scholar] [CrossRef]
- Gulcin, I. Antioxidant and antiradical activities of l-carnitine. Life Sci. 2006, 78, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Othman, A.; Ismail, A.; Ghani, A.N.; Adenan, I. Antioxidant capacity and phenolic content of cocoa beans. Food Chem. 2007, 100, 1523–1530. [Google Scholar] [CrossRef]
- Ou, B.; Huang, D.; Hampsch-Woodill, M.; Flanagan, J.A.; Deemer, E.K. Analysis of antioxidant activities of common vegetables employing oxygen radical absorbance capacity (ORAC) and ferric reducing antioxidant power (FRAP) assays: A comparative study. J. Agric. Food Chem. 2002, 50, 3122–3128. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Jang, H.D. Nrf2-mediated HO-1 induction coupled with the ERK signaling pathway contributes to indirect antioxidant capacity of caffeic acid phenethyl ester in HepG2 cells. Int. J. Mol. Sci. 2014, 15, 12149–12156. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Sinha, D.; Saha, P.P.; Marthala, H.; D’Silva, P. Magmas functions as a ROS regulator and provides cytoprotection against oxidative stress-mediated damages. Cell Death Dis. 2014, 5, e1394. [Google Scholar] [CrossRef] [PubMed]
- Zeevalk, G.D.; Manzino, L.; Sonsalla, P.K.; Bernard, L.P. Characterization of intracellular elevation of glutathione (GSH) with glutathione monoethyl ester and GSH in brain and neuronal cultures: Relevance to Parkinson’s disease. Exp. Neurol. 2007, 203, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Ndisang, J.F. A critical and comprehensive insight on heme oxygenase and related products including carbon monoxide, bilirubin, biliverdin and ferritin in type -1 and type-2 diabetes. Curr. Pharm. Des. 2014, 20, 1370–1391. [Google Scholar] [CrossRef]
- Zou, X.; Gao, J.; Zheng, Y.; Wang, X.; Chen, C.; Cao, K.; Xu, J.; Li, Y.; Lu, W.; Liu, J.; Feng, Z. Zeaxanthin induces Nrf2-mediated phase II enzymes in protection of cell death. Cell Death Dis. 2014, 5, e1218. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-kostova, A.T.; Wang, X.J. Induction of the Keap1/Nrf2/ARE pathway by oxidizable diphenol. Chem. Biol. Interact. 2008, 192, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Boettler, U.; Sommerfeld-Volz, N.; Pahlke, G.; Teller, N.; Somoza, V.; Lang, R.; Hofmann, T.; Marko, D. Coffee constituents as modulator of Nrf2 nuclear translocation and ARE (EpRE)-dependent gene expression. J. Nutr. Biochem. 2011, 22, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Han, Y.; Rong, J. Potential roles of PI3K/Akt and Nrf2-Keap1 pathways in regulating hormesis of Z-ligustilide in PC12 cells against oxygen and glucose deprivation. Neuropharmacology 2012, 62, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Choi, R.J.; Cheng, M.S.; Kim, Y.S. Desoxyrhapontigenin up-regulates Nrf2-mediated heme oxygenase-1 expression in macrophages and inflammatory lung injury. Redox Biol. 2014, 2, 504–512. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Lv, L.; Lu, Q.; Li, Y.; Zhang, F.; Lin, M.; Gao, D.; Liu, K.; Tian, X.; Yao, J. Gallic acid attenuates dimethylnitrosamine-induced acute liver injury in mice through Nrf2-mediated induction of heme oxygenase-1 and glutathione-s-transferase α 3. Med. Chem. 2014, 4, 663–669. [Google Scholar] [CrossRef]
- Kumar, N.; Bhandari, P.; Singh, B.; Gupta, A.P.; Kaul, V.K. Reversed phase-HPLC for rapid determination of polyphenols in flowers of rose species. J. Sep. Sci. 2008, 31, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, F.; Goto, K.; Seto, R.; Suzuki, M.; Sakai, M.; Hara, Y. Scavenging effects of tea catechins and their derivatives on 1,1-diphenyl-2-picrylhydrazyl radical. Free Radic. Biol. Med. 1996, 21, 895–902. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Kumar, A.; Chattopadhyay, S. DNA damage protecting activity and antioxidant potential of pudina extract. Food Chem. 2007, 100, 1377–1384. [Google Scholar] [CrossRef]
- Saeed, N.; Khan, M.R.; Shabbir, M. Antioxidant activity, total phenolic and total flavonoid contents of whole plant extracts Torilis leptophylla L. BMC Complement. Altern. Med. 2012, 12, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Apak, R.; Guclu, K.; Ozyurek, M.; Karademir, S.E. Novel total antioxidant capacity index for dietary polyphenols and vitamins C and E, using their cupric ion reducing capability in the presence of neocuproine: CUPRAC method. J. Agric. Food Chem. 2004, 52, 7970–7981. [Google Scholar] [CrossRef] [PubMed]
- Salgueiro, A.C.; Folmer, V.; da Rosa, H.S.; Costa, M.T.; Boligon, A.A.; Paula, F.R.; Roos, D.H.; Puntel, G.O. In vitro and in silico antioxidant and toxicological activities of Achyrocline satureioides. J. Ethnopharmacol. 2016, 194, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Bender, C.; Graziano, S.; Zimmerman, B.F.; Weidlich, H.H. Antioxidant potential of aqueous plant extracts assessed by the cellular antioxidant activity assay. Am. J. Biol. Life Sci. 2014, 2, 72–79. [Google Scholar]
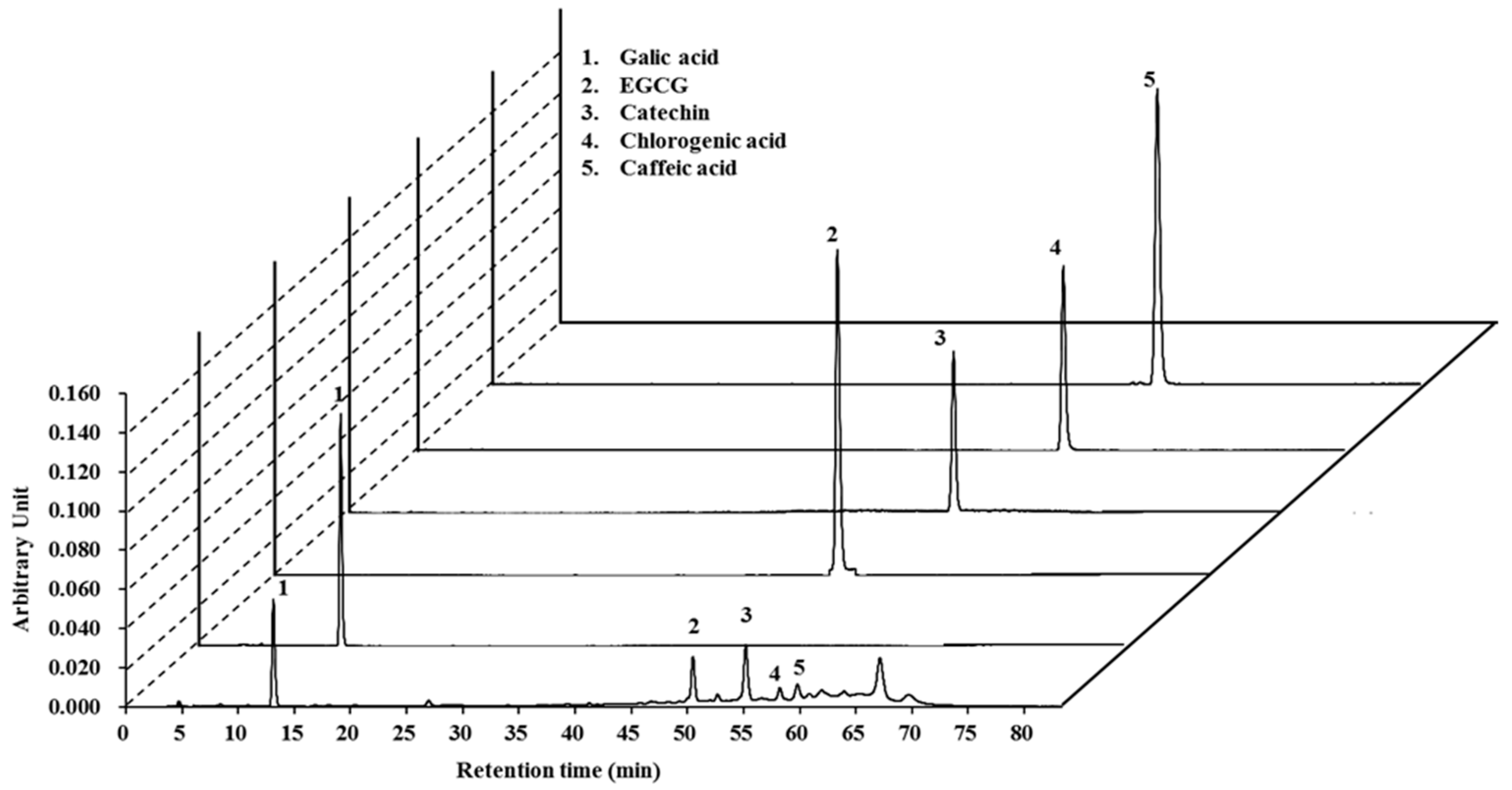

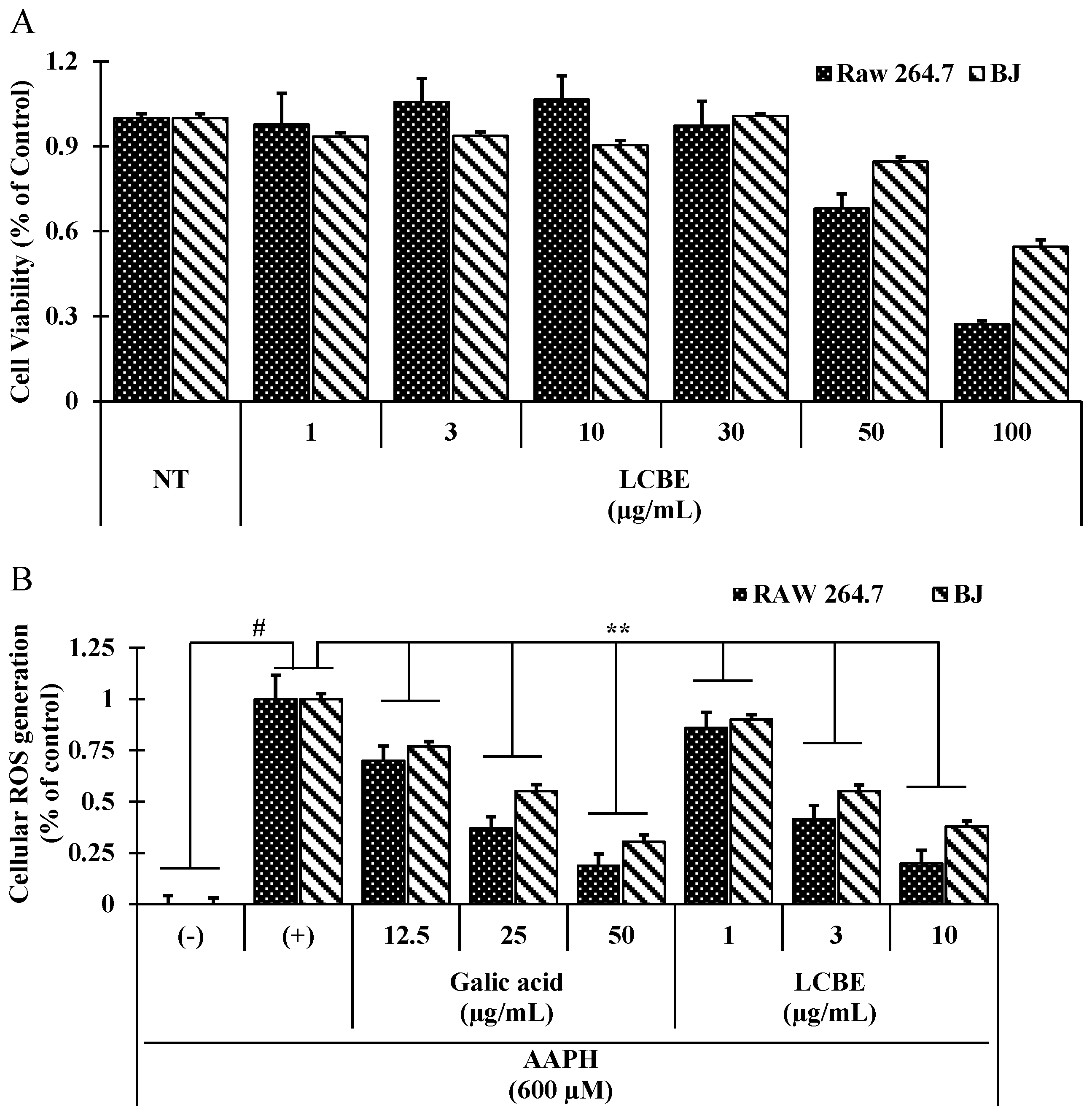
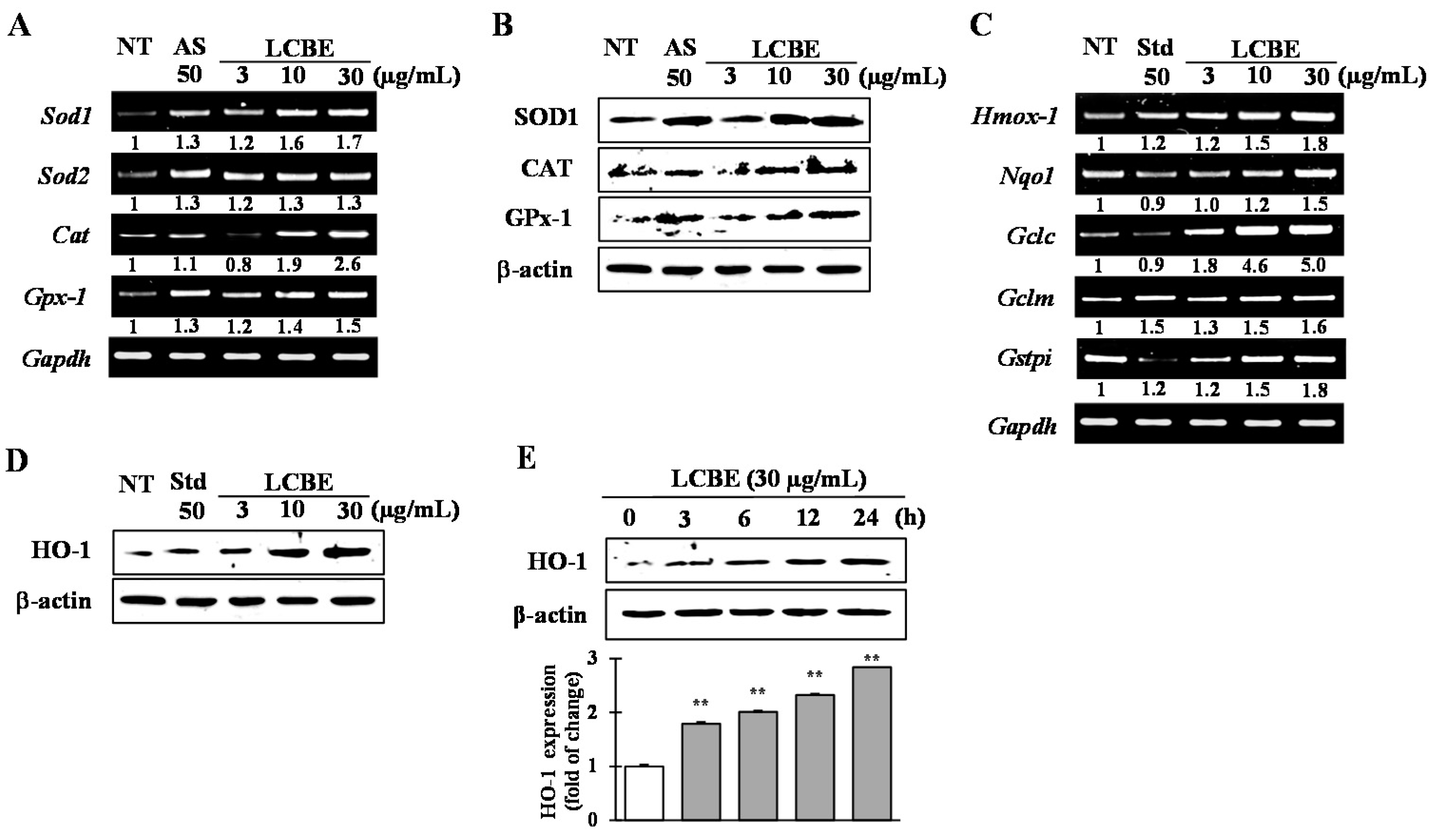
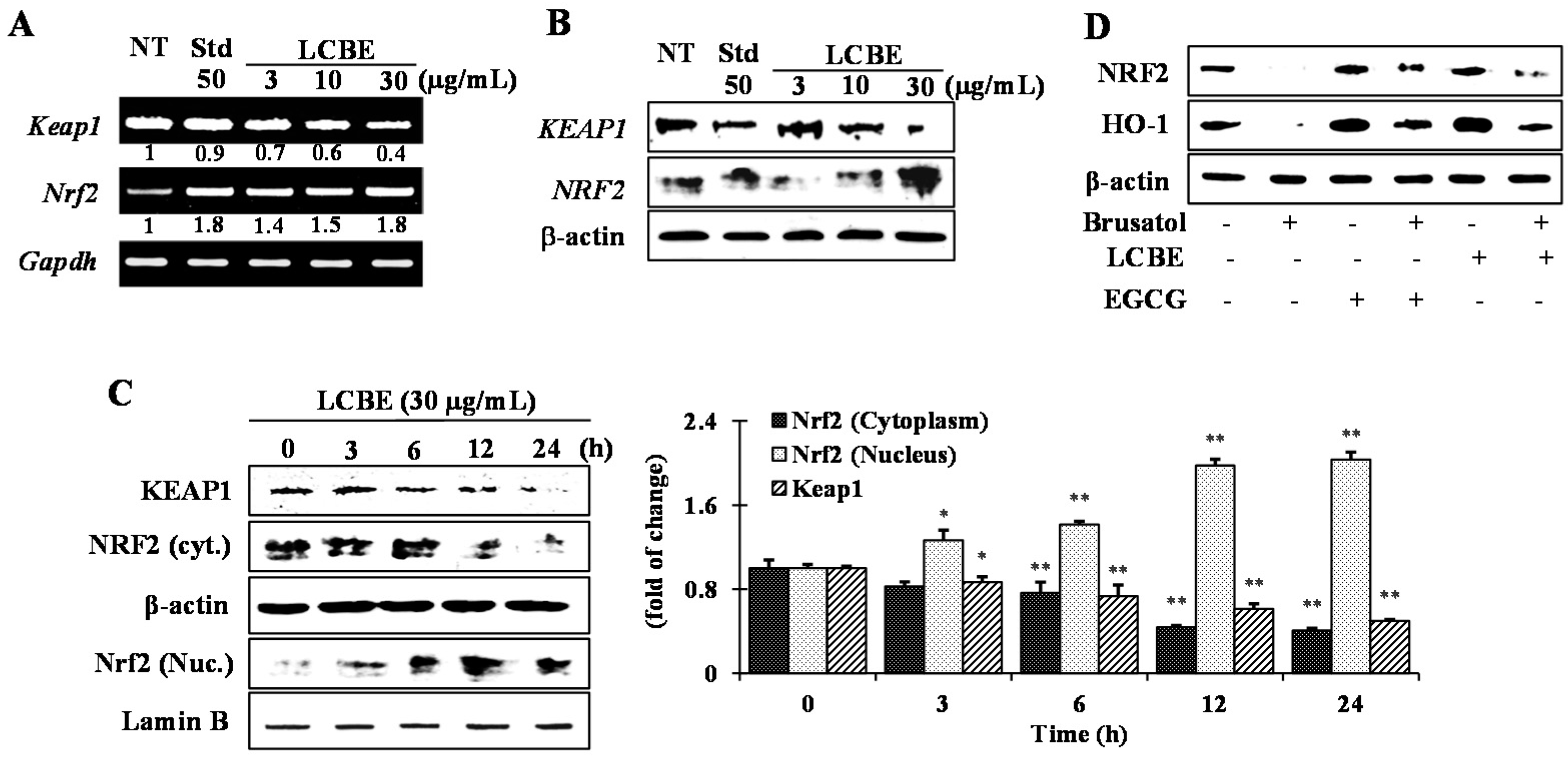
| ID Substances | Toxic Risk by ACD/Labs 1; admetSAR 2; pkCSM 3; PreADMET 4 | |||||
|---|---|---|---|---|---|---|
| Phytoconstituents | Mutagenic | Carcinogenic | Cardiotoxic | Skin Irritant | Reproductive System Toxicity 1 | Hepatotoxic |
| Gallic acid | N1, | N1, | N3, | N1 | N3 | |
| N2, | N2, | (+)2, | ||||
| N3, | N4, | N3, | ||||
| (+)4 | (+)4 | |||||
| EGCG | (+)1, | (+)1, | N3 | N1 | N3 | |
| N2, | N2, | (+)2, | ||||
| N3, | (+)4, | N3, | ||||
| (+)4 | (++)4 | |||||
| Cathechin | (+)1, | (+)1, | N3 | (+)1 | N3 | |
| N2, | N2, | (+)2, | ||||
| N3, | N4, | N3, | ||||
| (+)4 | (++)4 | |||||
| Chlorogenic acid | (+)1, | N1, | N3 | N1 | (+)3 | |
| N2, | N2, | (+)2, | ||||
| N3, | (+)4, | N3, | ||||
| N4 | (+++)4 | |||||
| Caffeic acid | (+)1, | N1, | N3 | N1 | N3 | |
| N2, | N2, | (+)2, | ||||
| N3, | (+)4, | N3, | ||||
| (+)4 | (++)4 | |||||
| Phytoconstituents | Main Predicted Properties by PASS Online | Pa # | Pi # |
|---|---|---|---|
| Gallic acid | Antiseptic | 0.921 | 0.003 |
| Astringent | 0.884 | 0.001 | |
| Antiinfective | 0.828 | 0.005 | |
| Fibrinolytic | 0.823 | 0.004 | |
| Mucomembranous protector | 0.814 | 0.015 | |
| Mucositis treatment | 0.798 | 0.014 | |
| Kidney function stimulant | 0.779 | 0.004 | |
| HO-1 expression enhancer | 0.732 | 0.005 | |
| EGCG | HMOX1 expression enhancer | 0.985 | 0.001 |
| Mucomembranous protector | 0.950 | 0.003 | |
| Fibrinolytic | 0.942 | 0.003 | |
| Free radical scavenger | 0.934 | 0.001 | |
| Astringent | 0.927 | 0.001 | |
| Antioxidant | 0.827 | 0.003 | |
| Cardioprotectant | 0.822 | 0.003 | |
| Hepatoprotectant | 0.820 | 0.004 | |
| Antihemorrhagic | 0.794 | 0.002 | |
| Antiviral (Influenza) | 0.771 | 0.003 | |
| Antihypercholesterolemic | 0.705 | 0.008 | |
| Catechin | HO-1 expression enhancer | 0.971 | 0.001 |
| Mucomembranous protector | 0.962 | 0.003 | |
| Fibrinolytic | 0.959 | 0.002 | |
| Antihemorrhagic | 0.926 | 0.002 | |
| Astringent | 0.921 | 0.001 | |
| Free radical scavenger | 0.850 | 0.002 | |
| Antioxidant | 0.828 | 0.003 | |
| Chemopreventive | 0.791 | 0.004 | |
| Hepatoprotectant | 0.769 | 0.005 | |
| Chlorogenic acid | Choleretic | 0.920 | 0.001 |
| Free radical scavenger | 0.856 | 0.002 | |
| Antioxidant | 0.809 | 0.003 | |
| Antieczematic | 0.808 | 0.017 | |
| Antineoplastic | 0.785 | 0.014 | |
| Mucomembranous protector | 0.752 | 0.034 | |
| Caffeic acid | Mucomembranous protector | 0.945 | 0.003 |
| Mucositis treatment | 0.873 | 0.008 | |
| HO-1 expression enhancer | 0.799 | 0.004 | |
| Antiseptic | 0.782 | 0.004 | |
| Vasoprotector | 0782 | 0.006 | |
| Fibrinolytic | 0.750 | 0.009 | |
| Cytoprotectant | 0.722 | 0.039 | |
| Antieczematic | 0.702 | 0.005 |
| Gene Name | Sequences | |
|---|---|---|
| Sod1 | Forward | AAGCGGTGAACCAGTTGTGT |
| Reverse | GCCAATGATGGAATGCTCTC | |
| Sod2 | Forward | AACTCAGGTCGCTCTTCAGC |
| Reverse | CTGTAAGCGACCTTGCTCCT | |
| Gpx1 | Forward | ACACCGAGATGAACGATCTG |
| Reverse | ATGTACTTGGGGTCGGTCAT | |
| Cat | Forward | CACCCACGATATCACCAGATAC |
| Reverse | GAAGACTCCAGAAGTCCCAGAC | |
| Hmox1 | Forward | ACGCATATACCCGCTACCTG |
| Reverse | TCCTCTGTCAGCATCACCTG | |
| Gclc | Forward | GGAGGCGATGTTCTTGAGAC |
| Reverse | GGGTGCTTGTTTATGGCTTC | |
| Gclm | Forward | AGTTGCACAGCTGGACTCTG |
| Reverse | TCGGGTCATTGTGAGTCAGT | |
| Nqo1 | Forward | CTGGCCCATTCAGAGAAGAC |
| Reverse | GTCTGCAGCTTCCAGCTTCT | |
| Nrf2 | Forward | ACATCCTTTGGAGGCAAGAC |
| Reverse | GGGAATGTCTCTGCCAAAAG | |
| Gstpi | Forward | GCCCAGATGGATATGGTGAA |
| Reverse | ATGGGACGGTTCACATGTTC | |
| Keap1 | Forward | GCTACAACCCCATGACCAAC |
| Reverse | GCGGAGTTAAGCCGGTTAGT | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alam, M.B.; Kwon, K.-R.; Lee, S.-H.; Lee, S.-H. Lannea coromandelica (Houtt.) Merr. Induces Heme Oxygenase 1 (HO-1) Expression and Reduces Oxidative Stress via the p38/c-Jun N-Terminal Kinase–Nuclear Factor Erythroid 2-Related Factor 2 (p38/JNK–NRF2)-Mediated Antioxidant Pathway. Int. J. Mol. Sci. 2017, 18, 266. https://doi.org/10.3390/ijms18020266
Alam MB, Kwon K-R, Lee S-H, Lee S-H. Lannea coromandelica (Houtt.) Merr. Induces Heme Oxygenase 1 (HO-1) Expression and Reduces Oxidative Stress via the p38/c-Jun N-Terminal Kinase–Nuclear Factor Erythroid 2-Related Factor 2 (p38/JNK–NRF2)-Mediated Antioxidant Pathway. International Journal of Molecular Sciences. 2017; 18(2):266. https://doi.org/10.3390/ijms18020266
Chicago/Turabian StyleAlam, Md Badrul, Kyoo-Ri Kwon, Seok-Hyun Lee, and Sang-Han Lee. 2017. "Lannea coromandelica (Houtt.) Merr. Induces Heme Oxygenase 1 (HO-1) Expression and Reduces Oxidative Stress via the p38/c-Jun N-Terminal Kinase–Nuclear Factor Erythroid 2-Related Factor 2 (p38/JNK–NRF2)-Mediated Antioxidant Pathway" International Journal of Molecular Sciences 18, no. 2: 266. https://doi.org/10.3390/ijms18020266







