Metabolomic Fingerprinting in the Comprehensive Study of Liver Changes Associated with Onion Supplementation in Hypercholesterolemic Wistar Rats
Abstract
:1. Introduction
2. Results and Discussion
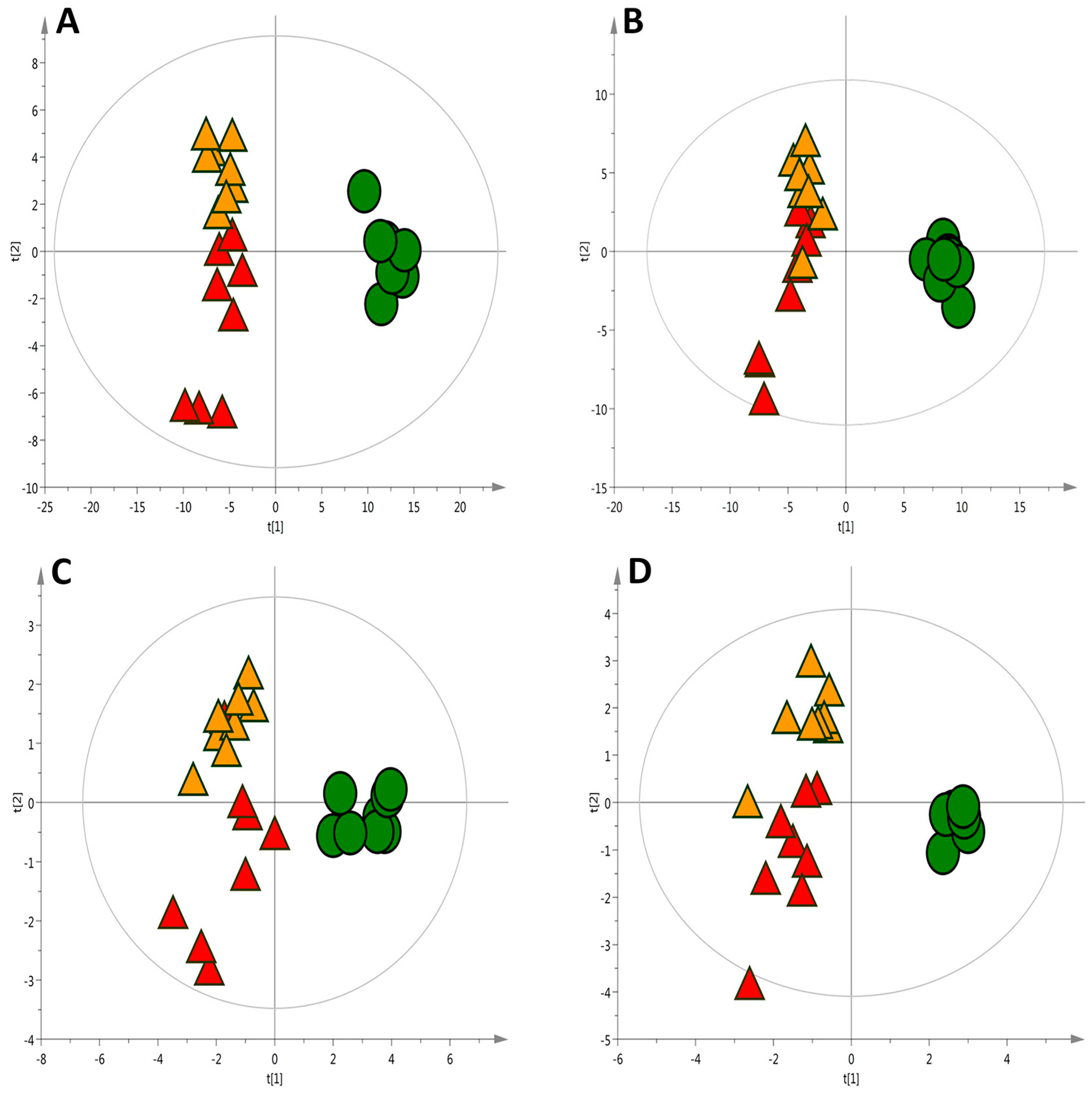
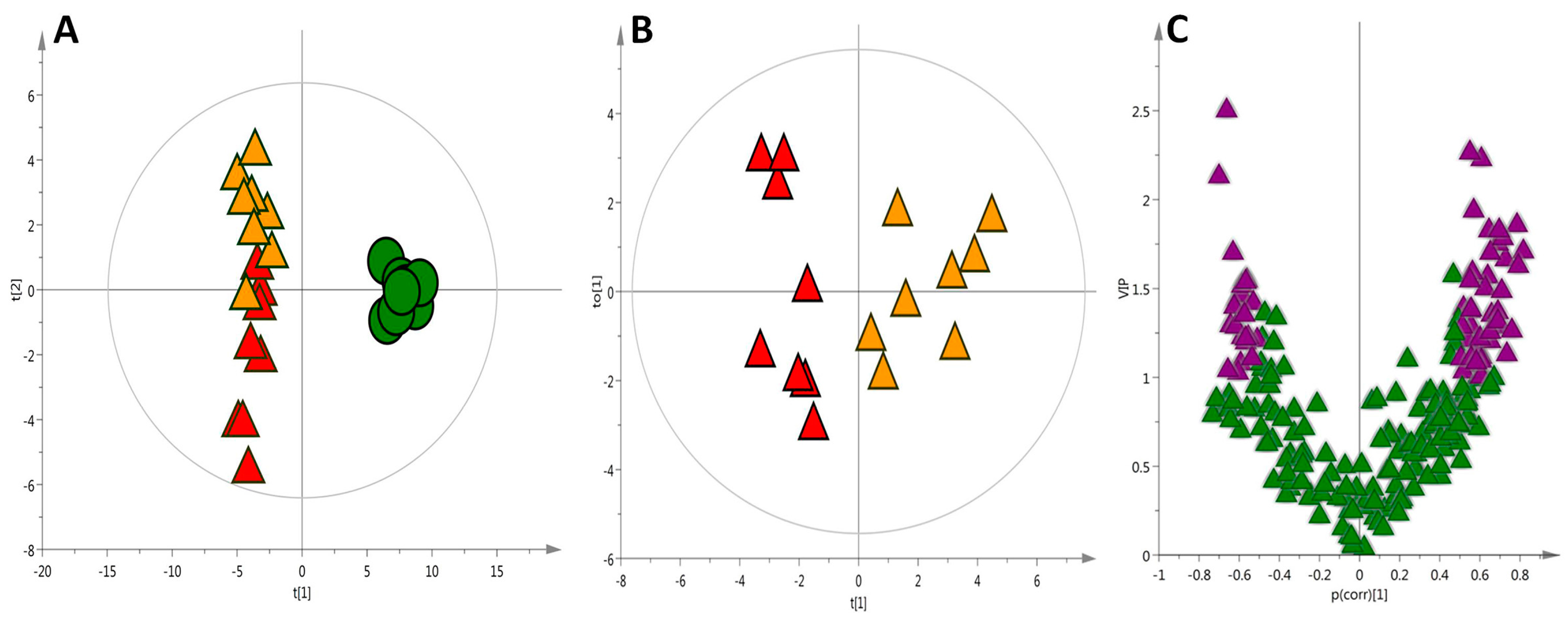
2.1. Liver Metabolic Fingerprinting by LC−MS
2.2. Liver Metabolic Fingerprinting by CE-MS
2.3. Liver Metabolic Fingerprinting by GC-MS
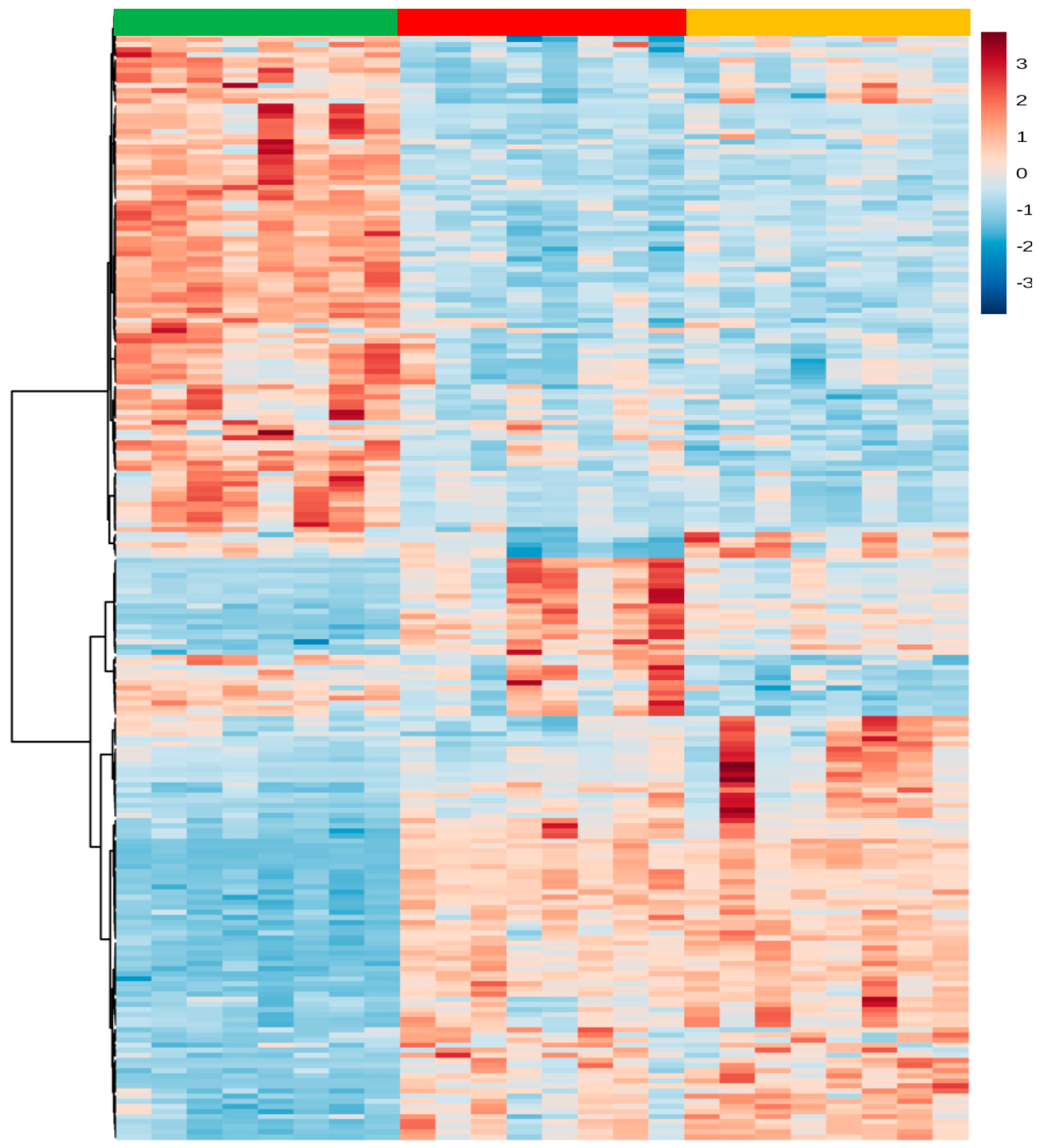
2.4. Global Metabolic Interpretation
3. Materials and Methods
3.1. Experimental Design and Diets
3.2. Liver Sampling and Homogenate Preparation
3.3. Metabolite Extraction
3.3.1. LC-MS Analysis
3.3.2. CE-MS Analysis
3.3.3. GC-MS Analysis
3.3.4. Randomization
3.4. QC Sample Preparation
3.5. Metabolic Fingerprinting
3.5.1. Liquid Chromatography-Quadrupole Time of Flight-Mass Spectrometry (LC-QTOF/MS Analysis)
3.5.2. Capillary Electrophoresis-Time of Flight-Mass Spectrometry (CE-TOF/MS Analysis)
3.5.3. Gas Chromatography-Quadrupole-Mass Spectrometry (GC-Q/MS Analysis)
3.5.4. Data Treatment
3.5.5. Statistical Analysis
3.5.6. Model Validation
3.5.7. Metabolite Identification
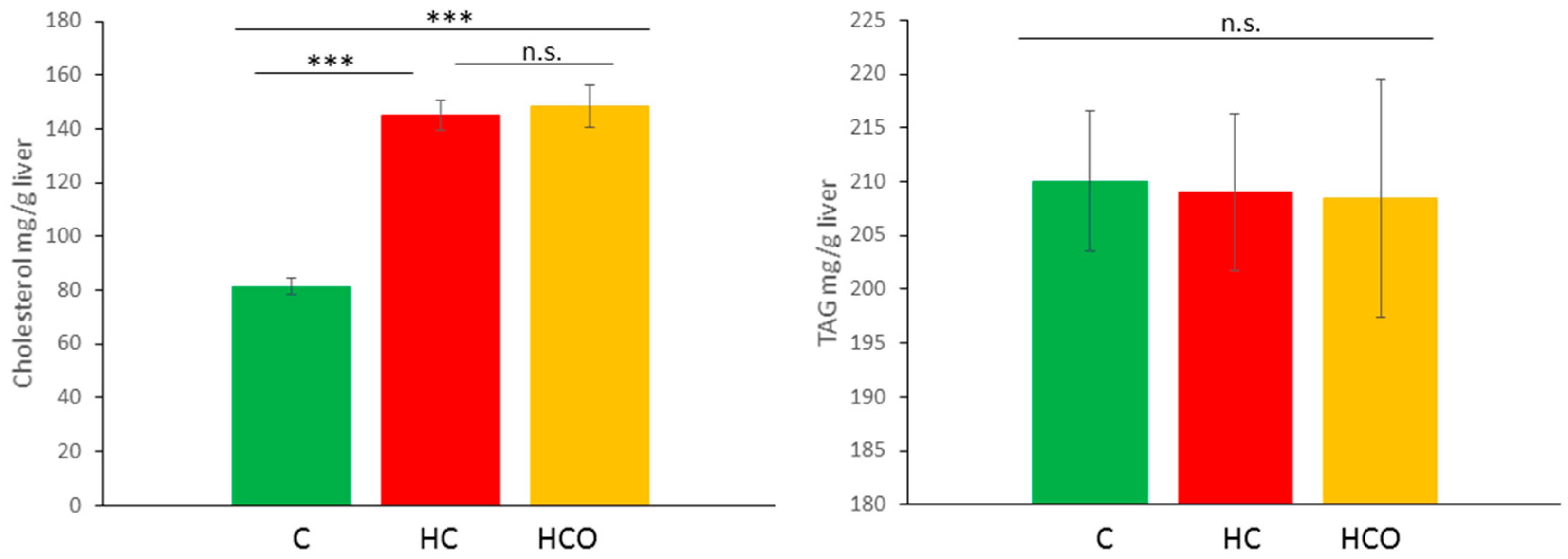
3.6. Determination of Cholesterol and TAG in Liver
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kim, E.J.; Kim, B.H.; Seo, H.S.; Lee, Y.J.; Kim, H.H.; Son, H.H.; Choi, M.H. Cholesterol-induced non-alcoholic fatty liver disease and atherosclerosis aggravated by systemic inflammation. PLoS ONE 2014, 9, e97841. [Google Scholar] [CrossRef] [PubMed]
- Bertot, L.C.; Adams, L.A. The natural course of non-alcoholic fatty liver disease. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Long, M.T.; Wang, N.; Larson, M.G.; Mitchell, G.F.; Palmisano, J.; Vasan, R.S.; Hoffmann, U.; Speliotes, E.K.; Vita, J.A.; Benjamin, E.J.; et al. Nonalcoholic fatty liver disease and vascular function: Cross-sectional analysis in the framingham heart study. Arteriorcler. Thromb. Vasc. Biol. 2015, 35, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- De Wit, N.J.; Afman, L.A.; Mensink, M.; Muller, M. Phenotyping the effect of diet on non-alcoholic fatty liver disease. J. Hepatol. 2012, 57, 1370–1373. [Google Scholar] [CrossRef] [PubMed]
- Salomone, F.; Godos, J.; Zelber-Sagi, S. Natural antioxidants for non-alcoholic fatty liver disease: Molecular targets and clinical perspectives. Liver Int. 2016, 36, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Suleria, H.A.R.; Butt, M.S.; Anjum, F.M.; Saeed, F.; Khalid, N. Onion: Nature protection against physiological threats. Crit. Rev. Food Sci. Nutr. 2015, 55, 50–66. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Thomson, M.; Afzal, M. Garlic and onions: Their effect on eicosanoid metabolism and its clinical relevance. Prostaglandins Leukot. Essent. Fat. Acids 2000, 62, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Ogunmodede, O.S.; Saalu, L.C.; Ogunlade, B.; Akunna, G.G.; Oyewopo, A.O. An evaluation of the hypoglycemic, antioxidant and hepatoprotective potentials of onion (Allium cepa L.) on alloxan-induced diabetic rabbits. Int. J. Pharm. 2012, 8, 21–29. [Google Scholar]
- Obioha, U.E.; Suru, S.M.; Ola-Mudathir, K.F.; Faremi, T.Y. Hepatoprotective potentials of onion and garlic extracts on cadmium-induced oxidative damage in rats. Biol. Trace Elem. Res. 2009, 129, 143–156. [Google Scholar] [CrossRef] [PubMed]
- Emamat, H.; Foroughi, F.; Eini-Zinab, H.; Taghizadeh, M.; Rismanchi, M.; Hekmatdoost, A. The effects of onion consumption on treatment of metabolic, histologic, and inflammatory features of nonalcoholic fatty liver disease. J. Diabetes Metab. Disord. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Roldan-Marin, E.; Sanchez-Moreno, C.; Lloria, R.; de Ancos, B.; Cano, M.P. Onion high-pressure processing: Flavonol content and antioxidant activity. LWT Food Sci. Technol. 2009, 42, 835–841. [Google Scholar] [CrossRef]
- González-Peña, D.; Colina-Coca, C.; Char, C.D.; Cano, M.P.; de Ancos, B.; Sánchez-Moreno, C. Hyaluronidase inhibiting activity and radical scavenging potential of flavonols in processed onion. J. Agric. Food Chem. 2013, 61, 4862–4872. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Wijeyesekera, A.; Taylor-Robinson, S.D.; Nicholson, J.K. The promise of metabolic phenotyping in gastroenterology and hepatology. Nat. Rev. Gastroenterol. Hepatol. 2015, 12, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Naz, S.; dos Santos, D.C.M.; Garcia, A.; Barbas, C. Analytical protocols based on LC-MS, GC-MS and CE-MS for nontargeted metabolomics of biological tissues. Bioanalysis 2014, 6, 1657–1677. [Google Scholar] [CrossRef] [PubMed]
- Beyoglu, D.; Idle, J.R. The metabolomic window into hepatobiliary disease. J. Hepatol. 2013, 59, 842–858. [Google Scholar] [CrossRef] [PubMed]
- Llorach, R.; Garcia-Aloy, M.; Tulipani, S.; Vazquez-Fresno, R.; Andres-Lacueva, C. Nutrimetabolomic strategies to develop new biomarkers of intake and health effects. J. Agric. Food Chem. 2012, 60, 8797–8808. [Google Scholar] [CrossRef] [PubMed]
- González-Peña, D.; Angulo, J.; Vallejo, S.; Colina-Coca, C.; de Ancos, B.; Sánchez-Ferrer, C.F.; Peiró, C.; Sánchez-Moreno, C. High-cholesterol diet enriched with onion affects endothelium-dependent relaxation and NADPH oxidase activity in mesenteric microvessels from Wistar rats. Nutr. Metab. 2014, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- González-Peña, D.; Dudzik, D.; Colina-Coca, C.; de Ancos, B.; García, A.; Barbas, C.; Sánchez-Moreno, C. Evaluation of onion as a functional ingredient in the prevention of metabolic impairments associated to diet-induced hypercholesterolaemia using a multiplatform approach based on LC-MS, CE-MS and GC-MS. J. Funct. Foods 2015, 19(Pt. A), 363–375. [Google Scholar] [CrossRef]
- González-Peña, D.; Checa, A.; de Ancos, B.; Wheelock, C.E.; Sánchez-Moreno, C. New insights into the effects of onion consumption on lipid mediators using a diet-induced model of hypercholesterolemia. Redox Biol. 2017, 11, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Gorden, D.L.; Ivanova, P.T.; Myers, D.S.; McIntyre, J.O.; VanSaun, M.N.; Wright, J.K.; Matrisian, L.M.; Brown, H.A. Increased diacylglycerols characterize hepatic lipid changes in progression of human nonalcoholic fatty liver disease; comparison to a murine model. PLoS ONE 2011, 6, e22775. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.M.; Zhang, B.; Xue, Y.; Li, Z.J.; Wang, J.F.; Xue, C.H.; Yanagita, T. The mechanism of dietary cholesterol effects on lipids metabolism in rats. Lipids Health Dis. 2010, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Moore, D.D.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef] [PubMed]
- Chitraju, C.; Trotzmuller, M.; Hartler, J.; Wolinski, H.; Thallinger, G.G.; Lass, A.; Zechner, R.; Zimmermann, R.; Kofeler, H.C.; Spener, F. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 2012, 53, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- González-Peña, D.; Dudzik, D.; Colina-Coca, C.; de Ancos, B.; Garcia, A.; Barbas, C.; Sánchez-Moreno, C. Multiplatform metabolomic fingerprinting as a tool for understanding hypercholesterolemia in Wistar rats. Eur. J. Nutr. 2016, 55, 997–1010. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Botolin, D.; Xu, J.; Christian, B.; Mitchell, E.; Jayaprakasam, B.; Nair, M.G.; Peters, J.M.; Busik, J.V.; Olson, L.K.; et al. Regulation of hepatic fatty acid elongase and desaturase expression in diabetes and obesity. J. Lipid Res. 2006, 47, 2028–2041. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, G.H. Evaluation of antioxidant and inhibitory activities for different subclasses flavonoids on enzymes for rheumatoid arthritis. J. Food Sci. 2010, 75, H212–H217. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.Q.; Li, Z.; Xie, W.R.; Liu, C.M.; Liu, S.S. Quercetin protects mouse liver against CCL4-induced inflammation by the TLR2/4 and MAPK/NF-κB pathway. Int. Immunopharmacol. 2015, 28, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Ro, J.Y.; Ryu, J.H.; Park, H.J.; Cho, H.J. Onion (Allium cepa L.) peel extract has anti-platelet effects in rat platelets. Springerplus 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Cho, I.; Ahn, J.; Jeon, T.I.; Ha, T.Y. Quercetin reduces high-fat diet-induced fat accumulation in the liver by regulating lipid metabolism genes. Phytother. Res. 2013, 27, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Zhang, Z.C.; Hassan, W.; Li, Y.; Liu, J.; Shang, J. Amelioration of free fatty acid-induced fatty liver by quercetin-3-O-β-d-glucuronide through modulation of peroxisome proliferator-activated receptor-alpha/sterol regulatory element-binding protein-1c signaling. Hepatol. Res. 2016, 46, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Regulska-Ilow, B.; Ilow, R. The influence of quercetin on fatty-acid content in selected organs of rats on diets with fresh and oxidized fat. Adv. Clin. Exp. Med. 2008, 17, 15–26. [Google Scholar]
- Pepin, E.; Guay, C.; Delghingaro-Augusto, V.; Joly, E.; Madiraju, S.R.; Prentki, M. Short-chain 3-hydroxyacyl-coa dehydrogenase is a negative regulator of insulin secretion in response to fuel and non-fuel stimuli in ins832/13 β-cells. J. Diabetes 2010, 2, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Hsu, J.W.; Jahoor, F.; Coraza, I.; Bain, J.R.; Stevens, R.D.; Iyer, D.; Nalini, R.; Ozer, K.; Hampe, C.S.; et al. Pathogenesis of A−β+ ketosis-prone diabetes. Diabetes 2013, 62, 912–922. [Google Scholar] [CrossRef] [PubMed]
- Mai, M.; Tönjes, A.; Kovacs, P.; Stumvoll, M.; Fiedler, G.M.; Leichtle, A.B. Serum levels of acylcarnitines are altered in prediabetic conditions. PLoS ONE 2013, 8, e82459. [Google Scholar] [CrossRef] [PubMed]
- Holecek, M.; Sispera, L. Effects of arginine supplementation on amino acid profiles in blood and tissues in fed and overnight-fasted rats. Nutrients 2016, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Corbin, K.D.; Zeisel, S.H. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr. Opin. Gastroenterol. 2012, 28, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Calderón-Santiago, M.; Priego-Capote, F.; Galache-Osuna, J.G.; Luque de Castro, M.D. Method based on GC-MS to study the influence of tricarboxylic acid cycle metabolites on cardiovascular risk factors. J. Pharm. Biomed. Anal. 2013, 74, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Mailloux, R.J.; Beriault, R.; Lemire, J.; Singh, R.; Chenier, D.R.; Hamel, R.D.; Appanna, V.D. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS ONE 2007, 2, e690. [Google Scholar] [CrossRef] [PubMed]
- Johari, N.Z.; Ismail, I.S.; Sulaiman, M.R.; Abas, F.; Shaari, K. Acute toxicity and metabolomics analysis of hypocholesterolemic effect of Mentha piperita aqueous extract in Wistar rats. Int. J. Appl. Res. Nat. Prod. 2015, 8, 1–11. [Google Scholar]
- Aziz, A.A.; Edwards, C.A.; Lean, M.E.; Crozier, A. Absorption and excretion of conjugated flavonols, including quercetin-4′-O-β-glucoside and isorhamnetin-4′-O-β-glucoside by human volunteers after the consumption of onions. Free Radic. Res. 1998, 29, 257–269. [Google Scholar] [CrossRef] [PubMed]
- De Vries, J.H.; Hollman, P.C.; Meyboom, S.; Buysman, M.N.; Zock, P.L.; van Staveren, W.A.; Katan, M.B. Plasma concentrations and urinary excretion of the antioxidant flavonols quercetin and kaempferol as biomarkers for dietary intake. Am. J. Clin. Nutr. 1998, 68, 60–65. [Google Scholar] [PubMed]
- Noroozi, M.; Burns, J.; Crozier, A.; Kelly, I.E.; Lean, M.E. Prediction of dietary flavonol consumption from fasting plasma concentration or urinary excretion. Eur. J. Clin. Nutr. 2000, 54, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Williamson, G. Comparison of the urinary excretion of quercetin glycosides from red onion and aglycone from dietary supplements in healthy subjects: A randomized, single-blinded, cross-over study. Food Funct. 2015, 6, 1443–1448. [Google Scholar] [CrossRef] [PubMed]
- Mullen, W.; Edwards, C.A.; Crozier, A. Absorption, excretion and metabolite profiling of methyl-, glucuronyl-, glucosyl- and sulpho-conjugates of quercetin in human plasma and urine after ingestion of onions. Br. J. Nutr. 2006, 96, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, J.; Kim, M.K. Onion flesh and onion peel enhance antioxidant status in aged rats. J. Nutr. Sci. Vitaminol. 2007, 53, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Minami, Y.; Ippoushi, K.; Terao, J. Lowering effects of onion intake on oxidative stress biomarkers in streptozotocin-induced diabetic rats. J. Clin. Biochem. Nutr. 2007, 40, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Ige, S.F.; Akhigbe, R.E. Common onion (Allium cepa) extract reverses cadmium-induced organ toxicity and dyslipidaemia via redox alteration in rats. Pathophysiology 2013, 20, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Reeves, P.G. Components of the AIN-93 diets as improvements in the AIN-76A diet. J. Nutr. 1997, 127, 838s–841s. [Google Scholar] [PubMed]
- Naz, S.; García, A.; Barbas, C. Multiplatform analytical methodology for metabolic fingerprinting of lung tissue. Anal. Chem. 2013, 85, 10941–10948. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; Velzen, E.J.J.; Duijnhoven, J.P.M.; Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
| Compound Name | p(corr) | VIP | Compound Name | p(corr) | VIP |
|---|---|---|---|---|---|
| Hydroxybutyrylcarnitine | −0.66 | 2.5 | Glycocholic acid | 0.50 | 1.33 |
| Glucopyranosyl-glucopyranosyl-glucose | 0.55 | 2.26 | PC(40:4) or PE(43:4) | 0.69 | 1.32 |
| Disaccharides (lactose) | 0.60 | 2.23 | LPC(32:0) or LPE(35:0) | −0.64 | 1.29 |
| Palmitoylcarnitine | −0.70 | 2.13 | Tricosanedioic acid | 0.55 | 1.29 |
| Methyl oleate | 0.57 | 1.94 | PC(33:3) or PE(36:3) | 0.51 | 1.29 |
| Methyl linolenate | 0.78 | 1.86 | PC(37:4) or PE(P-40:3) | 0.69 | 1.29 |
| Eicosadienoic acid | 0.65 | 1.83 | LPC(17:2) or LPE(20:2) | 0.76 | 1.26 |
| Cholenoic acid | 0.70 | 1.83 | PC(38:6) | 0.65 | 1.26 |
| Tetracosahexaenoic acid | 0.72 | 1.78 | PC(40:9) | 0.50 | 1.25 |
| Eicosenoic acid | 0.69 | 1.75 | Xanthine | −0.59 | 1.23 |
| Methyl palmitate | 0.82 | 1.71 | PC(38:4) or PE(38:4) | −0.55 | 1.23 |
| Docosatetraenoic acid | 0.65 | 1.7 | PC(37:4) or PE(40:4) | 0.60 | 1.22 |
| O-phosphocolamine | −0.63 | 1.7 | Succinic acid | 0.55 | 1.22 |
| Oleic acid | 0.73 | 1.67 | DAG(42:8) | −0.56 | 1.22 |
| Octadecatrienoic acid | 0.79 | 1.63 | SM(d44:1) | 0.54 | 1.22 |
| Glucose | 0.56 | 1.59 | Linolenoyl ethanolamide | 0.65 | 1.21 |
| Glycodeoxycholic acid | 0.50 | 1.58 | SM(33:1) | −0.57 | 1.2 |
| Docosapentaenoic acid | 0.64 | 1.57 | PC(36:6) | 0.50 | 1.19 |
| Sedoheptulose | 0.55 | 1.55 | Heneicosatrienoic acid 21:3 | 0.58 | 1.15 |
| Fumaric acid | −0.56 | 1.54 | Docosahexaenoic acid methyl ester | 0.74 | 1.13 |
| Glucosylceramide (d34:1) | −0.57 | 1.54 | Sugar alcohols C6H14O6 | 0.61 | 1.12 |
| Glucosylceramide (34:0) | −0.57 | 1.54 | PC(36:6) or PE(39:6) | 0.50 | 1.11 |
| Uracil | −0.59 | 1.52 | PC(42:10) | 0.55 | 1.11 |
| LPC(O-13:1) or LPE(16:1) | 0.71 | 1.49 | PG(36:4) | −0.54 | 1.11 |
| 3-Hydroxybutyric acid | −0.58 | 1.47 | PC(40:8) | 0.50 | 1.11 |
| Malic acid | −0.61 | 1.44 | Dihydroxycholesterol | 0.60 | 1.1 |
| SM(d40:1) | −0.53 | 1.42 | PE(38:7) | 0.58 | 1.08 |
| PC(P-38:4) | −0.62 | 1.4 | Trans-hydroxy-proline | −0.59 | 1.08 |
| Deoxyuridine | 0.52 | 1.39 | Tetrahydroxy-cholanoic acid | 0.58 | 1.07 |
| PC(36:5) | 0.56 | 1.39 | PC(40:6) | −0.65 | 1.05 |
| Eicosatrienoic acid | 0.55 | 1.38 | PC(33:4) or PE(36:4) | 0.52 | 1.04 |
| Arachidonic acid | 0.69 | 1.37 | Dehydrosqualene | −0.61 | 1.03 |
| TAG(54:8) | −0.58 | 1.35 | PC(40:5) | 0.61 | 1.01 |
| Hexadecenoic acid | 0.66 | 1.35 | PC(38:2) | 0.60 | 1.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Peña, D.; Dudzik, D.; García, A.; Ancos, B.D.; Barbas, C.; Sánchez-Moreno, C. Metabolomic Fingerprinting in the Comprehensive Study of Liver Changes Associated with Onion Supplementation in Hypercholesterolemic Wistar Rats. Int. J. Mol. Sci. 2017, 18, 267. https://doi.org/10.3390/ijms18020267
González-Peña D, Dudzik D, García A, Ancos BD, Barbas C, Sánchez-Moreno C. Metabolomic Fingerprinting in the Comprehensive Study of Liver Changes Associated with Onion Supplementation in Hypercholesterolemic Wistar Rats. International Journal of Molecular Sciences. 2017; 18(2):267. https://doi.org/10.3390/ijms18020267
Chicago/Turabian StyleGonzález-Peña, Diana, Danuta Dudzik, Antonia García, Begoña De Ancos, Coral Barbas, and Concepción Sánchez-Moreno. 2017. "Metabolomic Fingerprinting in the Comprehensive Study of Liver Changes Associated with Onion Supplementation in Hypercholesterolemic Wistar Rats" International Journal of Molecular Sciences 18, no. 2: 267. https://doi.org/10.3390/ijms18020267