Whole Exome Sequencing in Psoriasis Patients Contributes to Studies of Acitretin Treatment Difference
Abstract
:1. Introduction
2. Results
2.1. Clinical Features of the Psoriatic Patients in Difference Phase
2.2. Whole Exome Sequencing Analysis
2.3. Univariate Analysis of Thirty-Four Positive SNPs
2.4. Multivariable Logistic Regression Analysis of CRB2 rs1105223T>C, ANKLE1 rs11086065 A>G, ARHGEF3 rs3821414 T>C and SFRP4 rs1802073G>T
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. DNA Extraction
4.3. Whole Exome Sequencing
4.4. Sequenom MassArray Analysis
4.5. Data Statistics and Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boehncke, W.H.; Schon, M.P. Psoriasis. Lancet 2015, 386, 983–994. [Google Scholar] [CrossRef]
- Griffiths, C.E.; Barker, J.N. Pathogenesis and clinical features of psoriasis. Lancet 2007, 370, 263–271. [Google Scholar] [CrossRef]
- Ma, C.; Harskamp, C.T.; Armstrong, E.J.; Armstrong, A.W. The association between psoriasis and dyslipidaemia: A systematic review. Br. J. Dermatol. 2013, 168, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Grayson, M. Psoriasis. Nature 2012, 492, S49. [Google Scholar] [CrossRef] [PubMed]
- Bowcock, A.M. The genetics of psoriasis and autoimmunity. Ann. Rev. Genomics Hum. Genet. 2005, 6, 93–122. [Google Scholar] [CrossRef] [PubMed]
- Bowcock, A.M.; Krueger, J.G. Getting under the skin: The immunogenetics of psoriasis. Nat. Rev. Immunol. 2005, 5, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Nickoloff, B.J.; Qin, J.Z.; Nestle, F.O. Immunopathogenesis of psoriasis. Clin. Rev. Allergy Immunol. 2007, 33, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Dogra, S.; Yadav, S. Acitretin in psoriasis: An evolving scenario. Int. J. Dermatol. 2014, 53, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Cao, W.; Ma, H.; Feng, J.; Li, X.; Zhang, X. Acitretin exerted a greater influence on T-helper (Th)1 and Th17 than on Th2 cells in treatment of psoriasis vulgaris. J. Dermatol. 2012, 39, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Becherel, P.A.; Mossalayi, M.D.; LeGoff, L.; Frances, C.; Chosidow, O.; Debre, P.; Arock, M. Mechanism of anti-inflammatory action of retinoids on keratinocytes. Lancet 1994, 344, 1570–1571. [Google Scholar] [CrossRef]
- Ormerod, A.D.; Campalani, E.; Goodfield, M.J. British Association of Dermatologists guidelines on the efficacy and use of acitretin in dermatology. Br. J. Dermatol. 2010, 162, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Campalani, E.; Allen, M.H.; Fairhurst, D.; Young, H.S.; Mendonca, C.O.; Burden, A.D.; Griffiths, C.E.; Crook, M.A.; Barker, J.N.; Smith, C.H. Apolipoprotein E gene polymorphisms are associated with psoriasis but do not determine disease response to acitretin. Br. J. Dermatol. 2006, 154, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Young, H.S.; Summers, A.M.; Read, I.R.; Fairhurst, D.A.; Plant, D.J.; Campalani, E.; Smith, C.H.; Barker, J.N.; Detmar, M.J.; Brenchley, P.E.; et al. Interaction between genetic control of vascular endothelial growth factor production and retinoid responsiveness in psoriasis. J. Investig. Dermatol. 2006, 126, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Chandran, V. The genetics of psoriasis and psoriatic arthritis. Clin. Rev. Allergy Immunol. 2013, 44, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Tervaniemi, M.H.; Siitonen, H.A.; Soderhall, C.; Minhas, G.; Vuola, J.; Tiala, I.; Sormunen, R.; Samuelsson, L.; Suomela, S.; Kere, J.; et al. Centrosomal localization of the psoriasis candidate gene product, CCHCR1, supports a role in cytoskeletal organization. PLoS ONE 2012, 7, e49920. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Momota, Y.; Oikawa, A.; Shinkai, H. Psoriatic skin expresses the transcription factor Gli1: Possible contribution of decreased neurofibromin expression. Br. J. Dermatol. 2006, 154, 619–623. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Liu, Z.; Xu, Z.; Ke, F.; Zhang, L.; Zhu, H.; Lou, F.; Wang, H.; Fei, Y.; Shi, Y.L.; et al. Epigenetic downregulation of SFRP4 contributes to epidermal hyperplasia in psoriasis. J. Immunol. 2015, 194, 4185–4198. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Duffin, K.C.; Helms, C.; Ding, J.; Stuart, P.E.; Goldgar, D.; Gudjonsson, J.E.; Li, Y.; Tejasvi, T.; Feng, B.J.; et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 2009, 41, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Reischl, J.; Schwenke, S.; Beekman, J.M.; Mrowietz, U.; Sturzebecher, S.; Heubach, J.F. Increased expression of Wnt5a in psoriatic plaques. J. Investig. Dermatol. 2007, 127, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Thelu, J.; Rossio, P.; Favier, B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002, 2, 7. [Google Scholar] [CrossRef]
- Gudjonsson, J.E.; Johnston, A.; Stoll, S.W.; Riblett, M.B.; Xing, X.; Kochkodan, J.J.; Ding, J.; Nair, R.P.; Aphale, A.; Voorhees, J.J.; et al. Evidence for altered Wnt signaling in psoriatic skin. J. Investig. Dermatol. 2010, 130, 1849–1859. [Google Scholar] [CrossRef] [PubMed]
- Thelu, J.; Viallet, J.P.; Dhouailly, D. Differential expression pattern of the three Fringe genes is associated with epidermal differentiation. J. Investig. Dermatol. 1998, 111, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Snow, G.E.; Kasper, A.C.; Busch, A.M.; Schwarz, E.; Ewings, K.E.; Bee, T.; Spinella, M.J.; Dmitrovsky, E.; Freemantle, S.J. Wnt pathway reprogramming during human embryonal carcinoma differentiation and potential for therapeutic targeting. BMC Cancer 2009, 9, 383. [Google Scholar] [CrossRef] [PubMed]
- Chien, A.J.; Conrad, W.H.; Moon, R.T. A Wnt survival guide: From flies to human disease. J. Investig. Dermatol. 2009, 129, 1614–1627. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.; Andrews, P.W. Expression of Wnt and Notch pathway genes in a pluripotent human embryonal carcinoma cell line and embryonic stem cell. Acta Pathol. Microbiol. Immunol. Scand. 2003, 111, 197–211. [Google Scholar] [CrossRef]
- Carmon, K.S.; Loose, D.S. Secreted frizzled-related protein 4 regulates two Wnt7a signaling pathways and inhibits proliferation in endometrial cancer cells. Mol. Cancer Res. 2008, 6, 1017–1028. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Nakajima, K.; Kikuno, N.; Yamamura, S.; Kawakami, K.; Suehiro, Y.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Wnt antagonist gene polymorphisms and renal cancer. Cancer 2009, 115, 4488–4503. [Google Scholar] [CrossRef] [PubMed]
- Green, A.C.; Kocovski, P.; Jovic, T.; Walia, M.K.; Chandraratna, R.A.; Martin, T.J.; Baker, E.K.; Purton, L.E. Retinoic acid receptor signalling directly regulates osteoblast and adipocyte differentiation from mesenchymal progenitor cells. Exp. Cell Res. 2017, 350, 284–297. [Google Scholar] [CrossRef] [PubMed]
- Froeling, F.E.; Feig, C.; Chelala, C.; Dobson, R.; Mein, C.E.; Tuveson, D.A.; Clevers, H.; Hart, I.R.; Kocher, H.M. Retinoic acid-induced pancreatic stellate cell quiescence reduces paracrine Wnt-β-catenin signaling to slow tumor progression. Gastroenterology 2011, 141, 1486.e14–1497.e14. [Google Scholar] [CrossRef] [PubMed]
- Mitsuishi, Y.; Hasegawa, H.; Matsuo, A.; Araki, W.; Suzuki, T.; Tagami, S.; Okochi, M.; Takeda, M.; Roepman, R.; Nishimura, M. Human CRB2 inhibits gamma-secretase cleavage of amyloid precursor protein by binding to the presenilin complex. J. Biol. Chem. 2010, 285, 14920–14931. [Google Scholar] [CrossRef] [PubMed]
- Brachner, A.; Braun, J.; Ghodgaonkar, M.; Castor, D.; Zlopasa, L.; Ehrlich, V.; Jiricny, J.; Gotzmann, J.; Knasmuller, S.; Foisner, R. The endonuclease Ankle1 requires its LEM and GIY-YIG motifs for DNA cleavage in vivo. J. Cell Sci. 2012, 125, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Johar, A.S.; Mastronardi, C.; Rojas-Villarraga, A.; Patel, H.R.; Chuah, A.; Peng, K.; Higgins, A.; Milburn, P.; Palmer, S.; Silva-Lara, M.F.; et al. Novel and rare functional genomic variants in multiple autoimmune syndrome and Sjogren’s syndrome. J. Transl. Med. 2015, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Lawrenson, K.; Kar, S.; McCue, K.; Kuchenbaeker, K.; Michailidou, K.; Tyrer, J.; Beesley, J.; Ramus, S.J.; Li, Q.; Delgado, M.K.; et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun. 2016, 7, 12675. [Google Scholar] [CrossRef] [PubMed]
- D’Amato, L.; Dell’Aversana, C.; Conte, M.; Ciotta, A.; Scisciola, L.; Carissimo, A.; Nebbioso, A.; Altucci, L. ARHGEF3 controls HDACi-induced differentiation via RhoA-dependent pathways in acute myeloid leukemias. Epigenetics 2015, 10, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, J.; Wozel, G. The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasis. Dermatology 2005, 210, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, S.; Ziaugra, L.; Tabbaa, D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. In Current Protocols in Human Genetics 2009; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009. [Google Scholar]
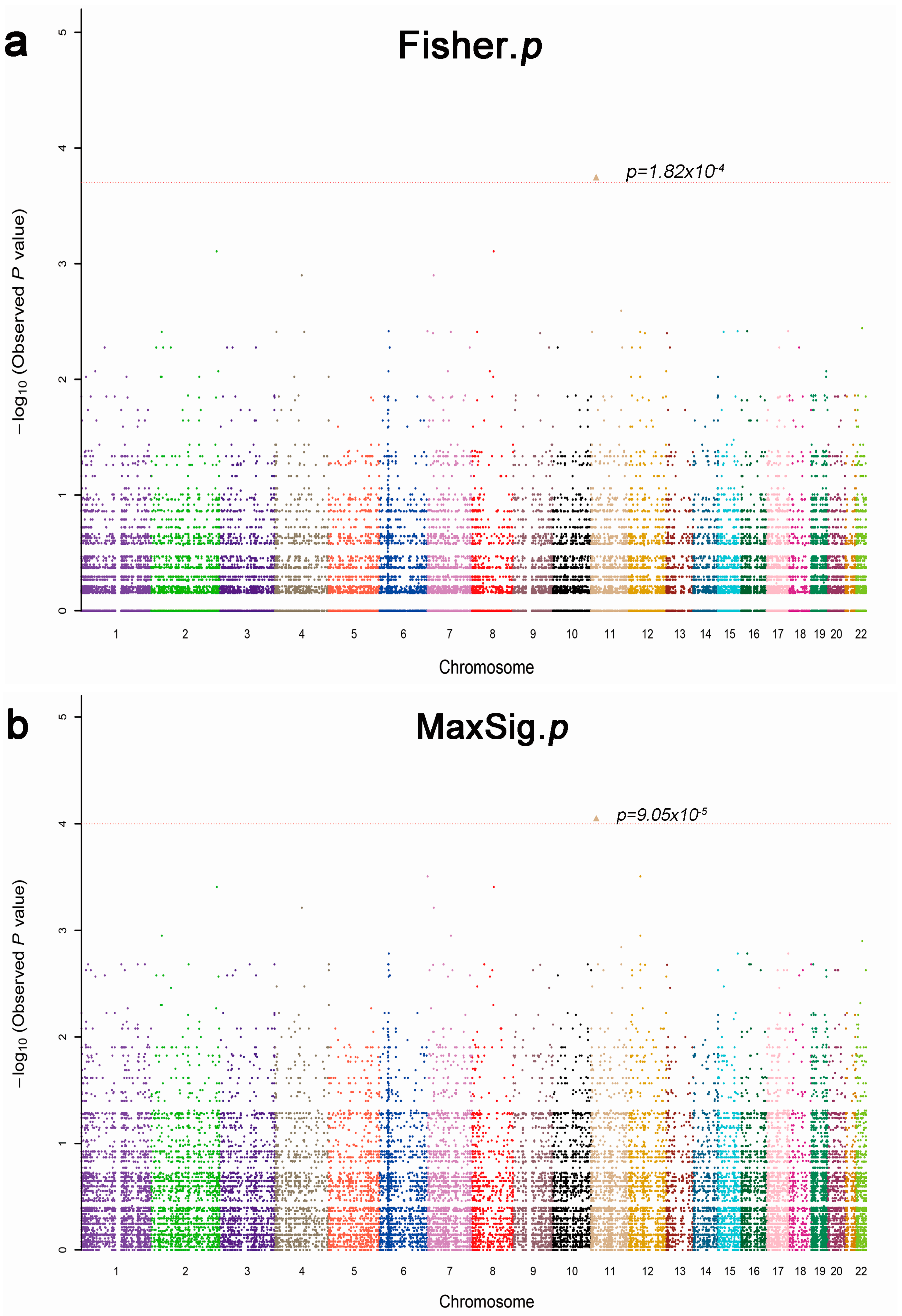
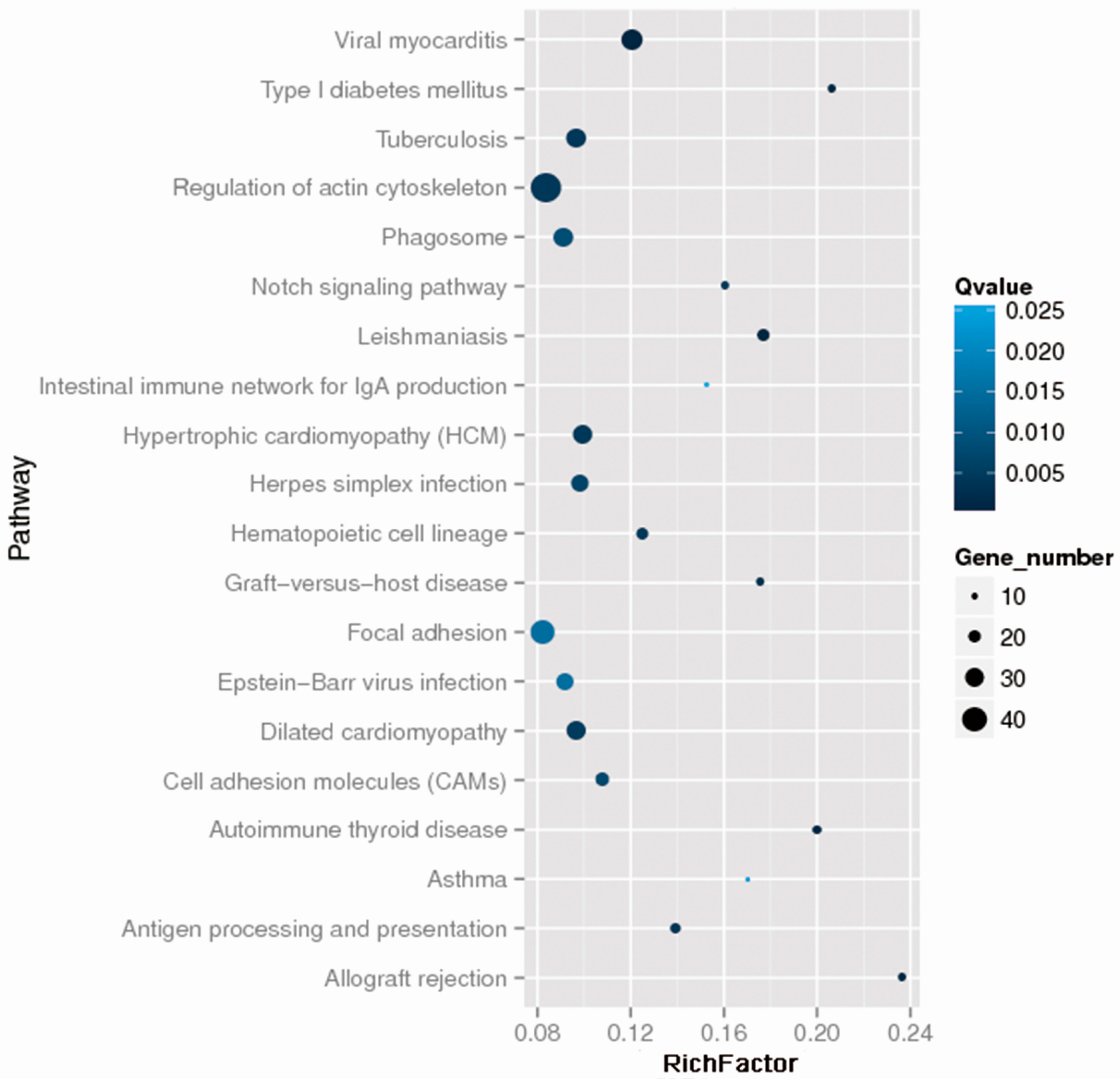
| Characteristics | Discovery Phase (n = 13) | p Value | Verification Phase (n = 166) | p Value | ||
|---|---|---|---|---|---|---|
| PASI < 75 | PASI ≥ 75 | PASI < 75 | PASI ≥ 75 | |||
| Age (mean ± SD) | 45 ± 12 | 52 ± 22 | 0.284 | 41 ± 13 | 43 ± 13 | 0.562 |
| Gender: Male, n (%) | 3 (37.5) | 4 (80) | 0.266 1 | 66 (66) | 52 (78.8) | 0.075 |
| Female, n (%) | 5 (62.5) | 1 (20) | 34 (34) | 14 (21.2) | ||
| BMI (mean ± SD) | 24.31 ± 4.44 | 23.25 ± 3.21 | 0.683 | 22.93 ± 3.90 | 23.08 ± 3.53 | 0.841 |
| Number | SNP ID | Gene | Chr | MAF | Location | Genotype | n | p for H–W | p Value (Drug Response) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Codominant | Dominant | Recessive | |||||||||
| 1 | rs10097933 | CSMD1 | 8 | 0.14 | Intron_variant | TT/CT/CC | 116/46/3 | 0.52 | 0.895 | 0.650 | 0.828 |
| 2 | rs10775247 | TICRR | 15 | 0.34 | Missense_variant | CC/CT/TT | 73/80/12 | 0.11 | 0.162 | 0.087 | 0.701 |
| 3 | rs1105223 | CRB2 | 9 | 0.47 | Missense_variant | TT/CT/CC | 63/64/29 | 0.08 | 0.015 | 0.497 | 0.020 |
| 4 | rs11086065 | ANKLE1 | 19 | 0.29 | Missense_variant | AA/AG/GG | 69/72/21 | 0.74 | 0.017 | 0.004 | 0.272 |
| 5 | rs1142825 | CALML3 | 10 | 0.49 | Synonymous_variant | AA/AG/GG | 61/83/22 | 0.45 | 0.919 | 0.680 | 0.906 |
| 6 | rs11674608 | CHRNG | 2 | 0.42 | Upstream_gene_variant | GG/CG/CC | 57/79/29 | 0.86 | 0.698 | 0.789 | 0.504 |
| 7 | rs13026692 | ALPP | 2 | 0.46 | Missense_variant | TT/AT/AA | 45/85/36 | 0.73 | 0.849 | 0.693 | 0.792 |
| 8 | rs1802073 | SFRP4 | 7 | 0.46 | Missense_variant | TT/GT/GG | 57/88/21 | 0.15 | 0.021 | 0.005 | 0.520 |
| 9 | rs2075333 | TSPAN11 | 12 | 0.22 | Missense_variant | CC/CT/TT | 75/78/13 | 0.24 | 0.878 | 0.794 | 0.624 |
| 10 | rs2076015 | TMX4 | 20 | 0.38 | Missense_variant | TT/CT/CC | 62/83/21 | 0.40 | 0.607 | 0.588 | 0.520 |
| 11 | rs2235638 | IFT140 | 16 | 0.17 | Missense_variant | GG/AG/AA | 97/59/10 | 0.80 | 0.514 | 0.269 | 0.515 |
| 12 | rs2241984 | PTPN5 | 11 | 0.22 | Intron_variant | GG/AG/AA | 114/46/6 | 0.62 | 0.938 | 0.818 | 0.743 |
| 13 | rs2303022 | ANXA6 | 5 | 0.48 | Intron_variant | GG/CG/CC | 39/97/30 | 0.03 | 0.531 | 0.573 | 0.427 |
| 14 | rs2303694 | ELL | 19 | 0.10 | Missense_variant | CC/CT/TT | 132/33/1 | 0.49 | 0.682 | 0.850 | 0.415 |
| 15 | rs2376558 | TPCN2 | 11 | 0.48 | Missense_variant | CC/CT/TT | 45/94/26 | 0.05 | 0.367 | 0.284 | 0.541 |
| 16 | rs2547065 | MUC16 | 19 | 0.21 | Missense_variant | GG/CG/CC | 107/55/4 | 0.32 | 0.790 | 0.857 | 0.541 |
| 17 | rs2933352 | MUC19 | 12 | 0.26 | Missense_variant | TT/CT/CC | 91/61/14 | 0.41 | 0.660 | 0.487 | 0.747 |
| 18 | rs2933353 | MUC19 | 12 | 0.31 | Missense_variant | CC/AC/AA | 74/73/19 | 0.88 | 0.875 | 0.615 | 0.782 |
| 19 | rs322118 | COL6A5 | 3 | 0.16 | Splice_region_variant | AA/AG/GG | 103/54/7 | 0.98 | 0.191 | 0.351 | 0.079 |
| 20 | rs335824 | NCBP2 | 3 | 0.24 | Upstream_gene_variant | TT/CT/CC | 91/67/8 | 0.33 | 0.259 | 0.562 | 0.178 |
| 21 | rs3733160 | TBCCD1 | 3 | 0.11 | Upstream_gene_variant | GG/AG/AA | 122/42/1 | 0.19 | 0.715 | 0.942 | 0.413 |
| 22 | rs3741595 | ORAI1 | 12 | 0.18 | Synonymous_variant | CC/CT/TT | 80/73/12 | 0.40 | 0.428 | 0.428 | 0.435 |
| 23 | rs3748664 | HHIPL2 | 1 | 0.50 | Synonymous_variant | CC/CG/GG | 42/89/34 | 0.30 | 0.397 | 0.868 | 0.181 |
| 24 | rs3817475 | GLI1 | 12 | 0.29 | Intron_variant | AA/AG/GG | 96/57/13 | 0.28 | 0.797 | 0.789 | 0.642 |
| 25 | rs3821414 | ARHGEF3 | 3 | 0.36 | 3_Prime_UTR_variant | TT/CT/CC | 69/73/24 | 0.51 | 0.006 | 0.002 | 0.044 |
| 26 | rs386624809 | SLC36A3 | 5 | 0.42 | Missense_variant | CC/CT/TT | 44/92/30 | 0.13 | 0.927 | 0.856 | 0.702 |
| 27 | rs47 | THSD7A | 7 | 0.25 | Missense_variant | CC/CT/TT | 82/77/6 | 0.02 | 0.373 | 0.923 | 0.164 |
| 28 | rs56310840 | GBA | 1 | 0.25 | Downstream_gene_variant | AA/AG/GG | 82/64/19 | 0.24 | 0.340 | 0.239 | 0.215 |
| 29 | rs7133914 | LRRK2 | 12 | 0.10 | Missense_variant | GG/AG/AA | 120/24/3 | 0.19 | 0.948 | 0.744 | 0.921 |
| 30 | rs7146310 | IPO4 | 14 | 0.40 | Missense_variant | GG/AG/AA | 60/76/30 | 0.49 | 0.592 | 0.479 | 0.659 |
| 31 | rs72927138 | LIN54 | 4 | 0.49 | 5_Prime_UTR_variant | GG/AG/AA | 45/74/42 | 0.31 | 0.360 | 0.449 | 0.398 |
| 32 | rs74976577 | ISYNA1 | 19 | 0.17 | Downstream_gene_variant | GG/GT/TT | 101/59/6 | 0.46 | 0.486 | 0.356 | 0.602 |
| 33 | rs76310711 | TNXB | 6 | - | Missense_variant | CC/CG/GG | 57/92/14 | 0.01 | 0.543 | 0.360 | 0.418 |
| 34 | rs916235 | C1QTNF6 | 22 | 0.45 | 3_Prime_UTR_variant | TT/CT/CC | 50/84/32 | 0.76 | 0.769 | 0.516 | 0.608 |
| Gene | SNPs | Genotypes/Alleles | PASI < 75 | PASI ≥ 75 | Adjusted OR 1 [95% CI] | p Value |
|---|---|---|---|---|---|---|
| n = 100 | n = 66 | |||||
| CRB2 | rs1105223 a | TT, n (%) | 40 (42.6) | 23 (37.1) | 1.00 | |
| CT, n (%) | 31 (33) | 33 (53.2) | 0.498 [0.234–1.062] | 0.071 | ||
| CC, n (%) | 23 (24.4) | 6 (9.7) | 1.852 [0.640–5.359] | 0.256 | ||
| CT/CC, n (%) | 54 (57.4) | 39 (62.9) | 0.734 [0.366–1.472] | 0.383 | ||
| TT/CT, n (%) | 71 (75.5) | 56 (90.3) | 0.371 [0.139–1.085] 2 | 0.048 | ||
| T, n (%) | 111 (59) | 79 (63.7) | 1.00 | |||
| C, n (%) | 77 (41) | 45 (36.3) | 1.118 [0.687–1.820] | 0.652 | ||
| ANKLE1 | rs11086065 b | AA, n (%) | 33 (33.7) | 36 (56.3) | 1.00 | |
| AG, n (%) | 50 (51) | 22 (34.3) | 2.922 [1.413–6.041] | 0.004 | ||
| GG, n (%) | 15 (15.3) | 6 (9.4) | 2.553 [0.851–7.652] | 0.094 | ||
| AG/GG, n (%) | 65 (66.3) | 28 (43.8) | 2.835 [1.436–5.600] | 0.003 | ||
| AA/AG, n (%) | 83 (84.7) | 58 (90.6) | 0.664 [0.235–1.875] 3 | 0.439 | ||
| A, n (%) | 116 (59.2) | 94 (73.4) | 1.00 | |||
| G, n (%) | 80 (40.8) | 34 (26.6) | 1.966 [1.181–3.271] | 0.009 | ||
| ARHGEF3 | rs3821414 | TT, n (%) | 51 (51) | 18 (27.3) | 1.00 | |
| CT, n (%) | 39 (39) | 34 (51.5) | 0.464 [0.222–0.968] | 0.041 | ||
| CC, n (%) | 10 (10) | 14 (21.2) | 0.256 [0.098–0.713] | 0.009 | ||
| CT/CC, n (%) | 49 (49) | 48 (72.7) | 0.402 [0.201–0.805] | 0.01 | ||
| TT/CT, n (%) | 90 (90) | 52 (78.8) | 2.465 [1.011–6.013] 4 | 0.047 | ||
| T, n (%) | 141 (70.5) | 70 (53) | 1.00 | |||
| C, n (%) | 59 (29.5) | 62 (47) | 0.497 [0.310–0.797] | 0.004 | ||
| SFRP4 | rs1802073 | GG, n (%) | 14 (14) | 7 (10.6) | 1.00 | |
| GT, n (%) | 60 (60) | 28 (42.4) | 1.196 [0.421–3.396] | 0.737 | ||
| TT, n (%) | 26 (26) | 31 (47) | 0.449 [0.153–1.321] | 0.146 | ||
| GT/TT, n (%) | 86 (86) | 59 (89.4) | 0.797 [0.296–2.149] | 0.655 | ||
| GG/GT, n (%) | 74 (74) | 35 (53) | 2.570 [1.294–5.107] 5 | 0.007 | ||
| G, n (%) | 88 (44) | 42 (31.8) | 1.00 | |||
| T, n (%) | 112 (56) | 90 (68.2) | 0.603 [0.374–0.971] | 0.037 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; He, Y.; Kuang, Y.; Li, J.; Zhang, J.; Chen, M.; Chen, W.; Su, J.; Zhao, S.; Liu, P.; et al. Whole Exome Sequencing in Psoriasis Patients Contributes to Studies of Acitretin Treatment Difference. Int. J. Mol. Sci. 2017, 18, 295. https://doi.org/10.3390/ijms18020295
Zhou X, He Y, Kuang Y, Li J, Zhang J, Chen M, Chen W, Su J, Zhao S, Liu P, et al. Whole Exome Sequencing in Psoriasis Patients Contributes to Studies of Acitretin Treatment Difference. International Journal of Molecular Sciences. 2017; 18(2):295. https://doi.org/10.3390/ijms18020295
Chicago/Turabian StyleZhou, Xingchen, Yijing He, Yehong Kuang, Jie Li, Jianglin Zhang, Mingliang Chen, Wangqing Chen, Juan Su, Shuang Zhao, Panpan Liu, and et al. 2017. "Whole Exome Sequencing in Psoriasis Patients Contributes to Studies of Acitretin Treatment Difference" International Journal of Molecular Sciences 18, no. 2: 295. https://doi.org/10.3390/ijms18020295





