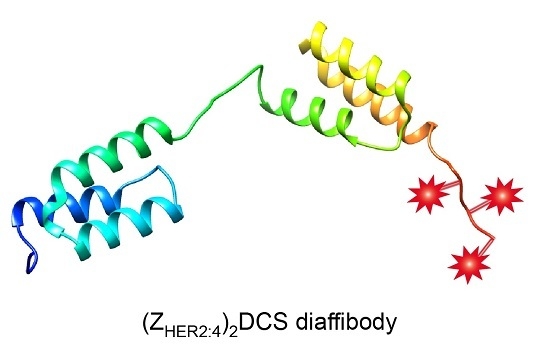
A Conjugate Based on Anti-HER2 Diaffibody and Auristatin E Targets HER2-Positive Cancer Cells
Abstract
:1. Introduction
2. Results
2.1. Expression and Purification of the (ZHER2:4)2DCS Diaffibody
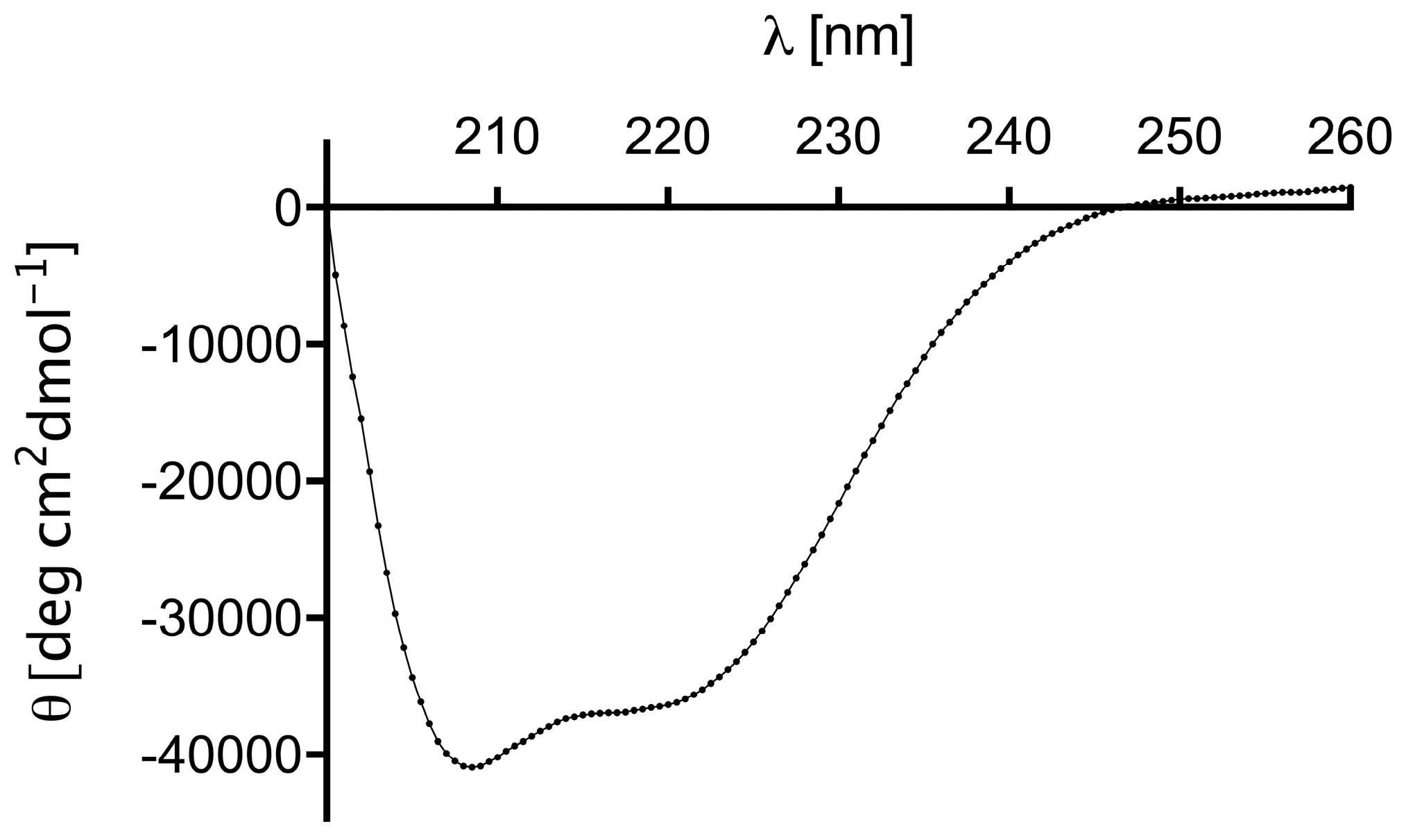
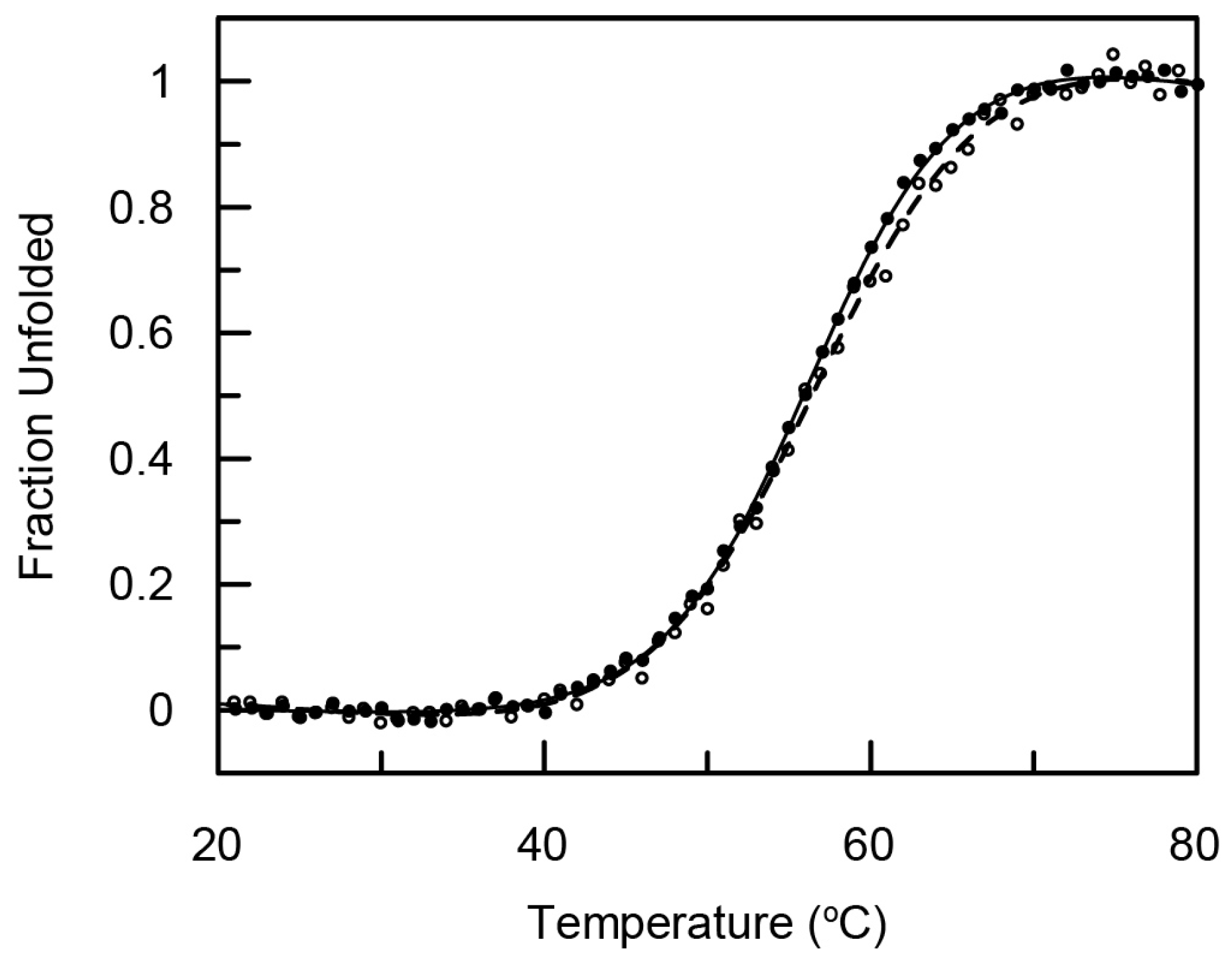
2.2. Structure and Thermal Stability of the (ZHER2:4)2DCS Diaffibody
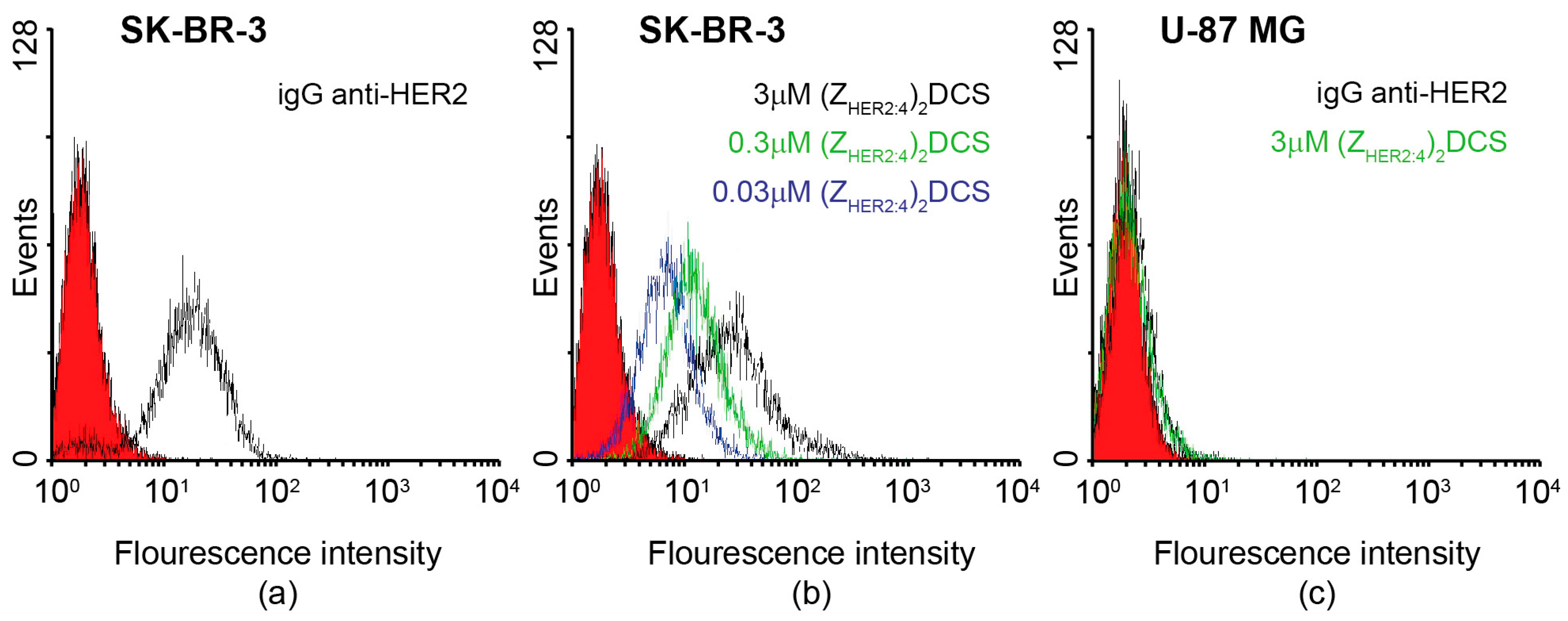
2.3. Specificity of the Dimeric Anti-HER2 Affibody
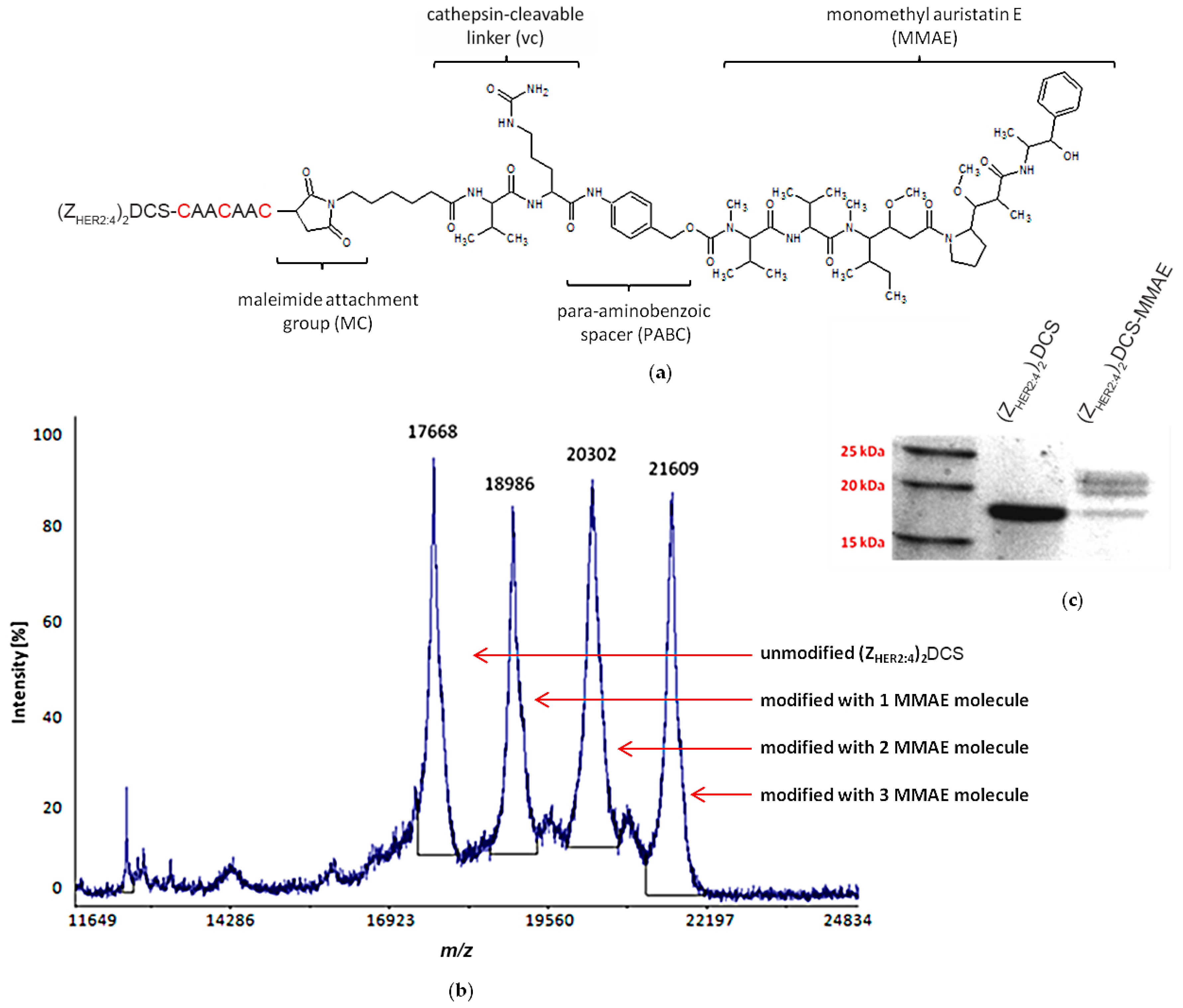
2.4. vcMMAE Conjugation and Conjugate Characterization
2.4.1. (ZHER2:4)2DCS-MMAE Preparation
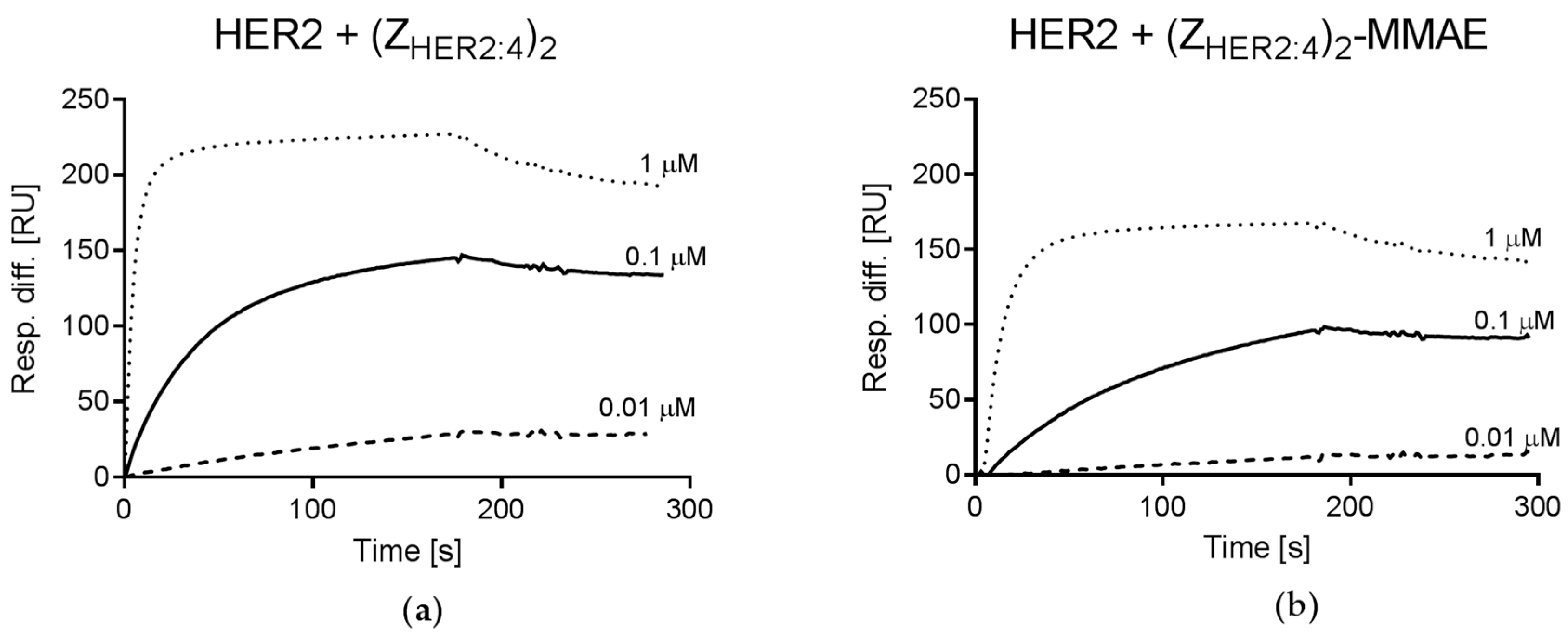
2.4.2. Analysis of (ZHER2:4)2DCS-MMAE Binding to HER2
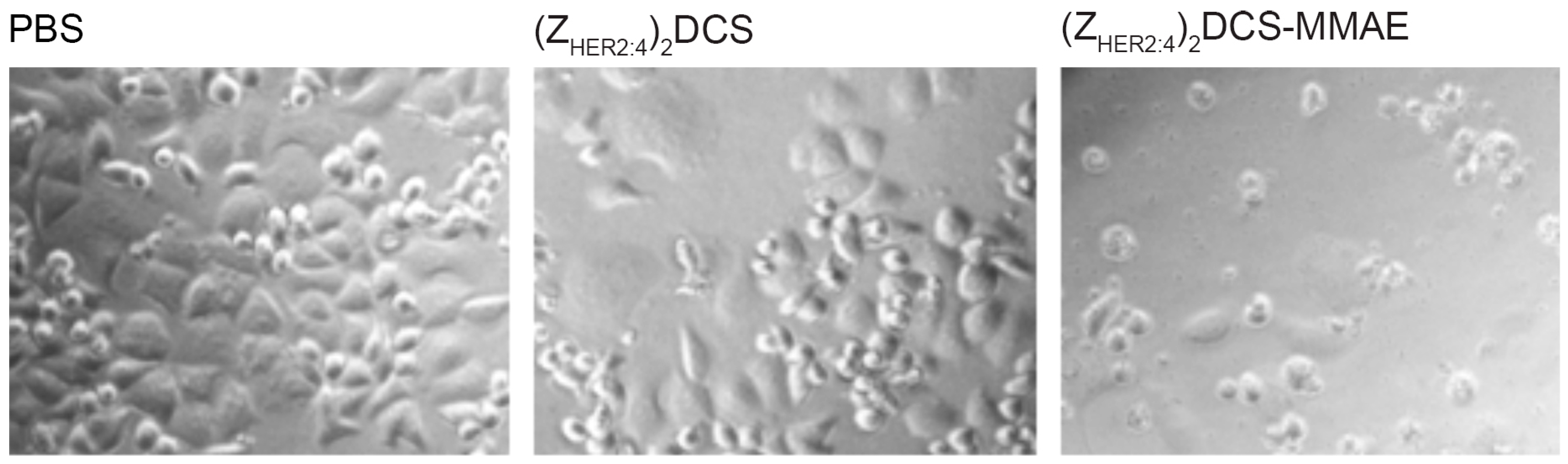
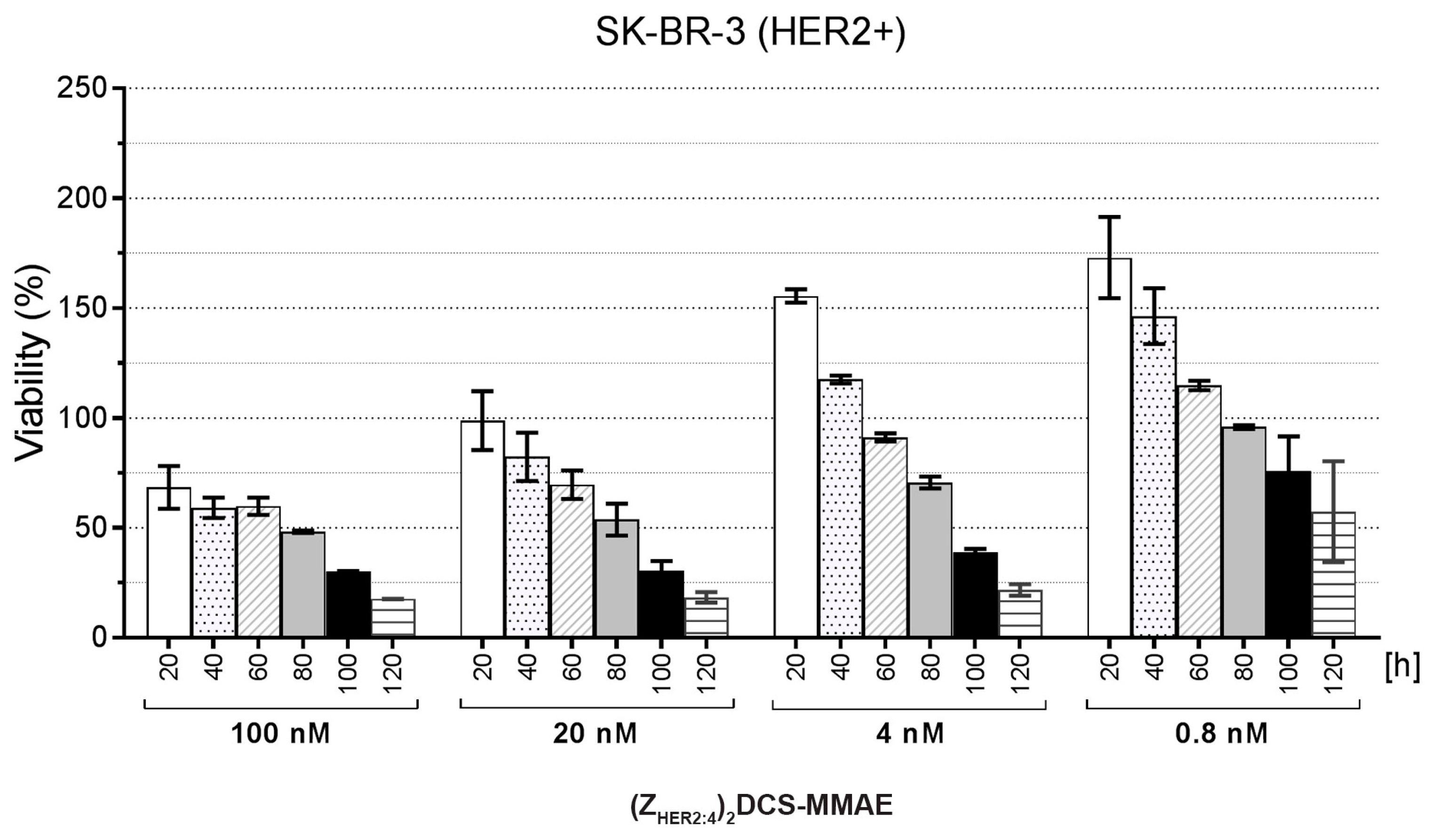
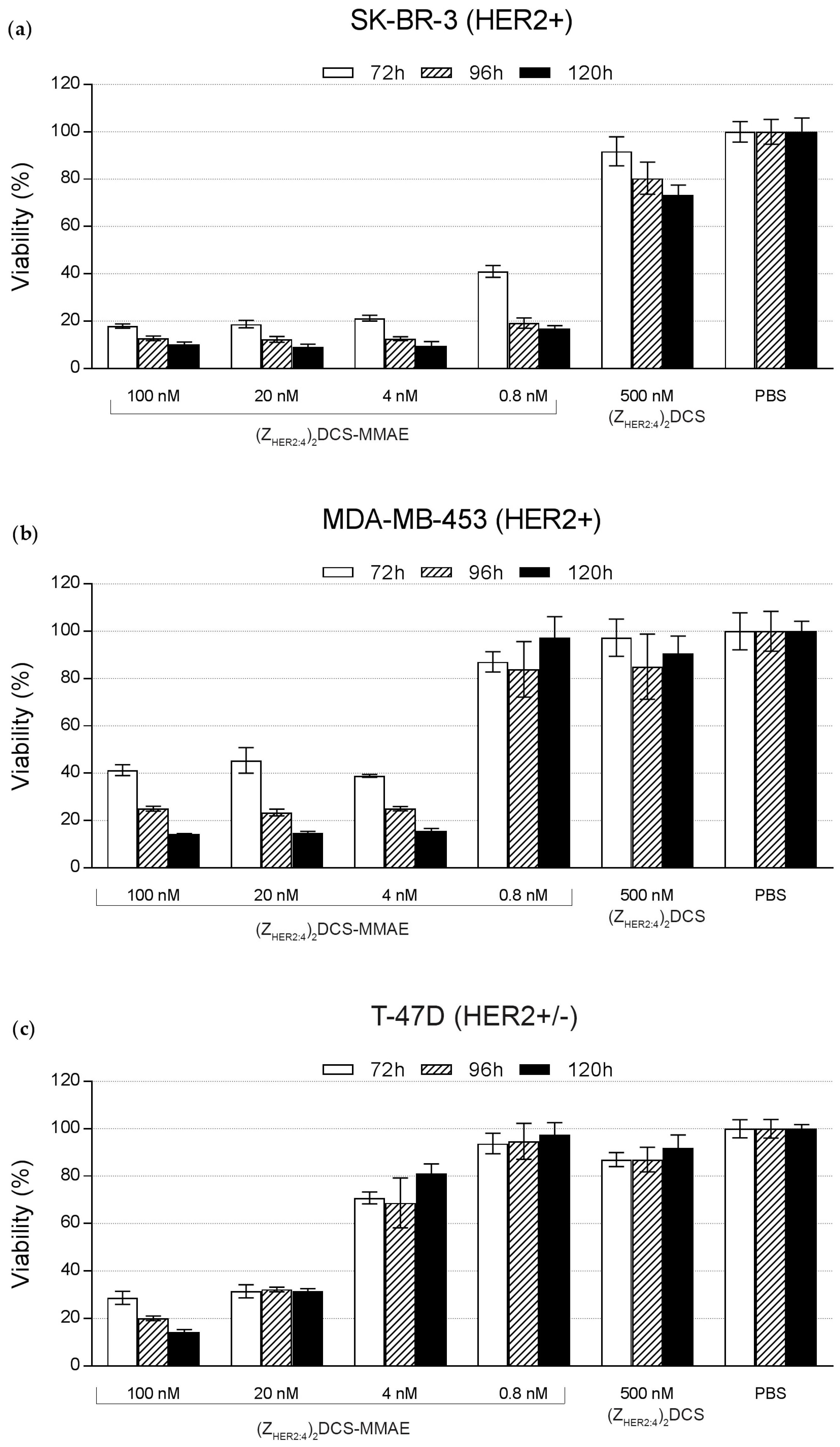
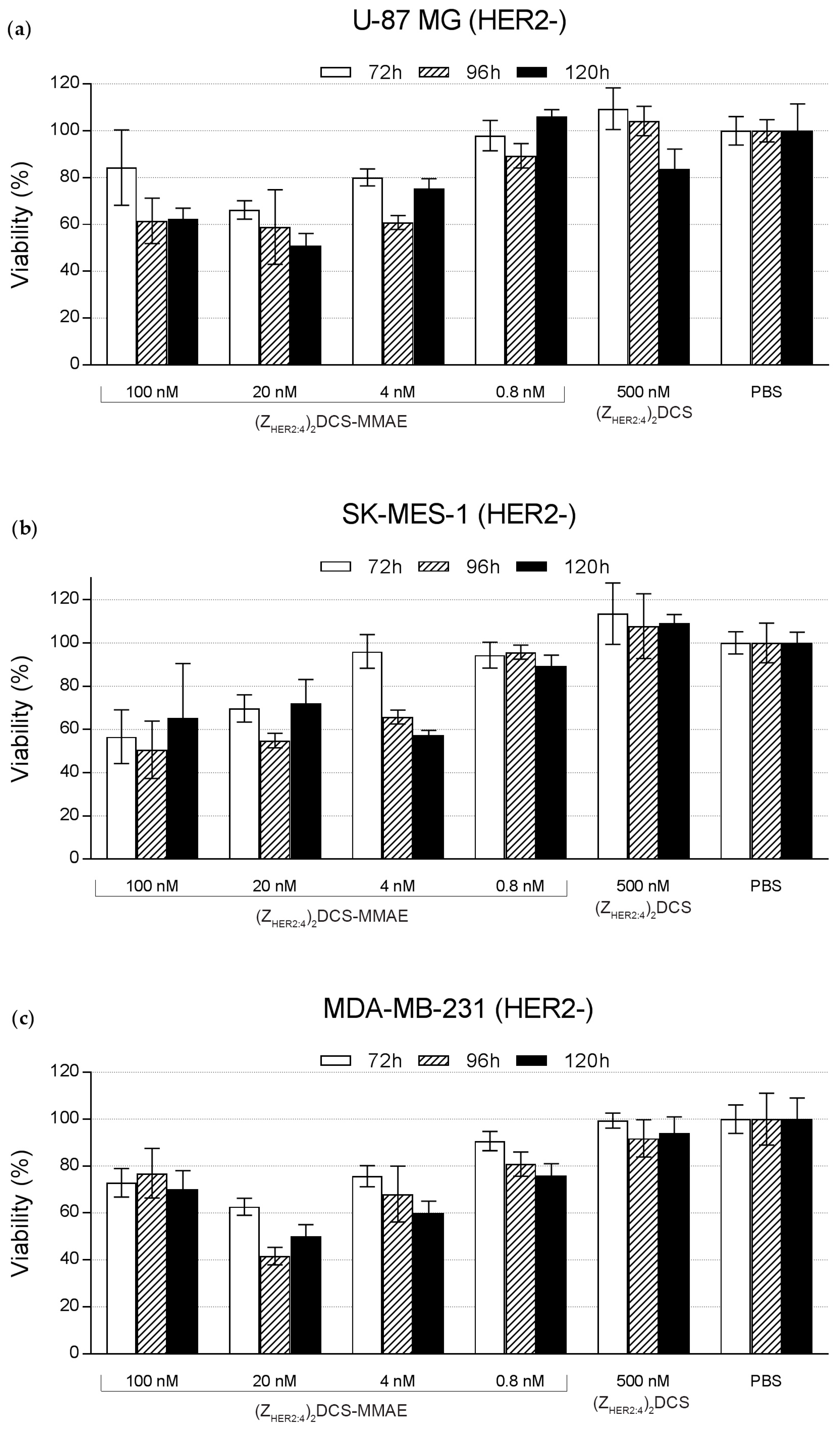
2.5. Cytotoxicity of the (ZHER2:4)2DCS-MMAE Conjugate
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Protein Expression and Purification
4.3. Circular Dichroism
4.4. Affinity Measurements
4.5. Fluorescein Labeling
4.6. Flow Cytometry
4.7. vcMMAE Conjugation
4.8. Mass Spectrometry
4.9. Cytotoxicity Measurements
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Kreitman, R.J. Immunotoxins for targeted cancer therapy. AAPS J. 2006, 18, E532–E551. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Chari, R.V.J. Antibody conjugate therapeutics: Challenges and potential. Clin. Cancer Res. 2011, 15, 6389–6397. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Teicher, B.A.; Hassan, R. Antibody-drug conjugates for cancer therapy. Lancet Oncol. 2016, 17, e254–e262. [Google Scholar] [CrossRef]
- Sochaj, A.M.; Świderska, K.W.; Otlewski, J. Current methods for the synthesis of homogeneous antibody-drug conjugates. Biotechnol. Adv. 2015, 1, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Teicher, B.A.; Doroshow, J.H. The promise of antibody-drug conjugates. N. Engl. J. Med. 2012, 8, 1847–1848. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, Y.V.; Goldmacher, V.S. Cell killing by antibody-drug conjugates. Cancer Lett. 2007, 255, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Senter, P.D.; Sievers, E.L. The discovery and development of brentuximab vedotin for use in relapsed Hodgkin lymphoma and systemic anaplastic large cell lymphoma. Nat. Biotechnol. 2012, 30, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Pan, B.; O’Connor, O.A. Brentuximab vedotin. Clin. Cancer Res. 2013, 19, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Niculescu-Duvaz, I. Trastuzumab emtansine, an antibody-drug conjugate for the treatment of HER2+ metastatic breast cancer. Curr. Opin. Mol. Ther. 2010, 12, 350–360. [Google Scholar] [PubMed]
- Peddi, P.F.; Hurvitz, S.A. Trastuzumab emtansine: The first targeted chemotherapy for treatment of breast cancer. Future Oncol. 2013, 9, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Saber, H.; Leighton, J.K. An FDA oncology analysis of antibody-drug conjugates. Regul. Toxicol. Pharmacol. 2015, 71, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.P.; Weiner, L.M. Monoclonal antibody therapy of cancer. Nat. Biotechnol. 2005, 23, 1147–1157. [Google Scholar] [CrossRef]
- Chames, P.; van Regenmortel, M.; Weiss, E.; Baty, D. Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharmacol. 2009, 2, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Casi, G.; Neri, D. Antibody-drug conjugates and small molecule-drug conjugates: Opportunities and challenges for the development of selective anticancer cytotoxic agents. J. Med. Chem. 2015, 25, 8751–8761. [Google Scholar] [CrossRef] [PubMed]
- Junutula, J.R.; Bhakta, S.; Raab, H.; Ervin, K.E.; Eigenbrot, C.; Vandlen, R.; Scheller, R.H.; Lowman, H.B. Rapid identification of reactive cysteine residues for site-specific labeling of antibody-Fabs. J. Immunol. Methods 2008, 20, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.L.; Reichert, J.M. Development trends for therapeutic antibody fragments. Nat. Biotechnol. 2009, 27, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Holliger, P.; Prospero, T.; Winter, G. “Diabodies”: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. USA 1993, 15, 6444–6448. [Google Scholar] [CrossRef]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 21, 423–426. [Google Scholar] [CrossRef]
- Cortez-Retamozo, V.; Backmann, N.; Senter, P.D.; Wernery, U.; de Baetselier, P.; Muyldermans, S.; Revets, H. Efficient cancer therapy with a nanobody-based conjugate. Cancer Res. 2004, 15, 2853–2857. [Google Scholar] [CrossRef]
- Oliveira, S.; Heukers, R.; Sornkom, J.; Kok, R.J.; van Bergen En Henegouwen, P.M. Targeting tumors with nanobodies for cancer imaging and therapy. J. Control. Release 2013, 28, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A. Alternative non-antibody scaffolds for molecular recognition. Curr. Opin. Biotechnol. 2007, 18, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Koide, A.; Abbatiello, S.; Rothgery, L.; Koide, S. Probing protein conformational changes in living cells by using designer binding proteins: Application to the estrogen receptor. Proc. Natl. Acad. Sci. USA 2002, 5, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A. Lipocalins as a scaffold. Biochim. Biophys. Acta 2000, 18, 337–350. [Google Scholar] [CrossRef]
- Stumpp, M.T.; Binz, H.K.; Amstutz, P. DARPins: A new generation of protein therapeutics. Drug Discov. Today 2008, 13, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Nord, K.; Gunneriusson, E.; Ringdahl, J.; Ståhl, S.; Uhlén, M.; Nygren, P.A. Binding proteins selected from combinatorial libraries of an α-helical bacterial receptor domain. Nat. Biotechnol. 1997, 15, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Wurch, T.; Pierré, A.; Depil, S. Novel protein scaffolds as emerging therapeutic proteins: From discovery to clinical proof-of-concept. Trends Biotechnol. 2012, 30, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Wikman, M.; Steffen, A.C.; Gunneriusson, E.; Tolmachev, V.; Adams, G.P.; Carlsson, J.; Ståhl, S. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 2004, 17, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Löfblom, J.; Feldwisch, J.; Tolmachev, V.; Carlsson, J.; Ståhl, S.; Frejd, F.Y. Affibody molecules: Engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett. 2010, 18, 2670–2680. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Lane, H.A.; Hynes, N.E. The role of overexpressed HER2 in transformation. Ann. Oncol. 2001, 12 (Suppl. 1), S9–S13. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; Mahato, R.; Cheng, K. The role of HER2 in cancer therapy and targeted drug delivery. J. Control. Release 2010, 15, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 9, 177–182. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, X.; Wang, S.; Wang, N.; Han, J.; Jia, L.; Ren, X. Monitoring therapeutic response of human ovarian cancer to trastuzumab by SPECT imaging with 99mTc-peptide-ZHER2:342. Nucl. Med. Biol. 2015, 42, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Sörensen, J.; Velikyan, I.; Sandberg, D.; Wennborg, A.; Feldwisch, J.; Tolmachev, V.; Orlova, A.; Sandström, M.; Lubberink, M.; Olofsson, H.; et al. measuring her2-receptor expression in metastatic breast cancer using [68Ga]ABY-025 Affibody PET/CT. Theranostics 2016, 1, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Hassan, M.; Fisher, R.; Chertov, O.; Chernomordik, V.; Kramer-Marek, G.; Gandjbakhche, A.; Capala, J. Affibody molecules for in vivo characterization of HER2-positive tumors by near-infrared imaging. Clin. Cancer Res. 2008, 15, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.C.; Wikman, M.; Tolmachev, V.; Adams, G.P.; Nilsson, F.Y.; Ståhl, S.; Carlsson, J. In vitro characterization of a bivalent anti-HER-2 affibody with potential for radionuclide-based diagnostics. Cancer Biother. Radiopharm. 2005, 20, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Steffen, A.C.; Orlova, A.; Wikman, M.; Nilsson, F.Y.; Ståhl, S.; Adams, G.P.; Tolmachev, V.; Carlsson, J. Affibody-mediated tumour targeting of HER-2 expressing xenografts in mice. Eur. J. Nucl. Med. Mol. Imaging 2006, 33, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. A self-consistent method for the analysis of protein secondary structure from circular dichroism. Anal. Biochem. 1993, 15, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Sreerama, N.; Woody, R.W. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004, 383, 318–351. [Google Scholar] [PubMed]
- Zheng, D.; Aramini, J.M.; Montelione, G.T. Validation of helical tilt angles in the solution NMR structure of the Z domain of Staphylococcal protein A by combined analysis of residual dipolar coupling and NOE data. Protein Sci. 2004, 13, 549–554. [Google Scholar] [CrossRef] [PubMed]
- Sochaj-Gregorczyk, A.M.; Serwotka-Suszczak, A.M.; Otlewski, J. A novel Affibody-auristatin E conjugate with a potent and selective activity against HER2+ cell lines. J. Immunother. 2016, 39, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Holliday, D.L.; Speirs, V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011, 13, 215. [Google Scholar] [CrossRef] [PubMed]
- Löfblom, J.; Frejd, F.Y.; Ståhl, S. Non-immunoglobulin based protein scaffolds. Curr. Opin. Biotechnol. 2011, 22, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Koduvayur, S.P.; Su, Y.; Kay, B.K.; Lavie, A. Targeted delivery of deoxycytidine kinase to HER2-positive cells enhances the efficacy of the nucleoside analog fludarabine. PLoS ONE 2016, 11, e157114. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Seijsing, J.; Frejd, F.Y.; Tolmachev, V.; Gräslund, T. Target-specific cytotoxic effects on HER2-expressing cells by the tripartite fusion toxin ZHER2:2891-ABD-PE38X8, including a targeting affibody molecule and a half-life extension domain. Int. J. Oncol. 2015, 47, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Zielinski, R.; Lyakhov, I.; Hassan, M.; Kuban, M.; Shafer-Weaver, K.; Gandjbakhche, A.; Capala, J. HER2-affitoxin: A potent therapeutic agent for the treatment of HER2-overexpressing tumors. Clin. Cancer Res. 2011, 1, 5071–5081. [Google Scholar] [CrossRef] [PubMed]
- Sörensen, J.; Sandberg, D.; Sandström, M.; Wennborg, A.; Feldwisch, J.; Tolmachev, V.; Åström, G.; Lubberink, M.; Garske-Román, U.; Carlsson, J.; et al. First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule. J. Nucl. Med. 2014, 5, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Honarvar, H.; Westerlund, K.; Altai, M.; Sandström, M.; Orlova, A.; Tolmachev, V.; Karlström, A.E. Feasibility of Affibody molecule-based PNA-mediated radionuclide pretargeting of malignant tumors. Theranostics 2016, 1, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Hynes, N.E.; Lane, H.A. ERBB receptors and cancer: The complexity of targeted inhibitors. Nat. Rev. Cancer 2005, 5, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Witton, C.J.; Reeves, J.R.; Going, J.J.; Cooke, T.G.; Bartlett, J.M. Expression of the HER1–4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 2003, 200, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Serwotka-Suszczak, A.M. The Conjugates of HER2 and Integrin αvβ3 Binding Proteins for Use in Anti-Cancer Therapies. Ph.D. Thesis, University of Wrocław, Wrocław, Poland, 13 April 2016. [Google Scholar]
- Ekerljung, L.; Lindborg, M.; Gedda, L.; Frejd, F.Y.; Carlsson, J.; Lennartsson, J. Dimeric HER2-specific affibody molecules inhibit proliferation of the SKBR-3 breast cancer cell line. Biochem. Biophys. Res. Commun. 2008, 377, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Doronina, S.O.; Toki, B.E.; Torgov, M.Y.; Mendelsohn, B.A.; Cerveny, C.G.; Chace, D.F.; DeBlanc, R.L.; Gearing, R.P.; Bovee, T.D.; Siegall, C.B.; et al. Development of potent monoclonal antibody auristatin conjugates for cancer therapy. Nat. Biotechnol. 2003, 7, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Francisco, J.A.; Cerveny, C.G.; Meyer, D.L.; Mixan, B.J.; Klussman, K.; Chace, D.F.; Rejniak, S.X.; Gordon, K.A.; DeBlanc, R.; Toki, B.E.; et al. cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood 2003, 4, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, M.; Krowarsch, D.; Wiedlocha, A.; Olsnes, S.; Otlewski, J. Highly stable mutants of human fibroblast growth factor-1 exhibit prolonged biological action. J. Mol. Biol. 2005, 352, 860–875. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Serwotka-Suszczak, A.M.; Sochaj-Gregorczyk, A.M.; Pieczykolan, J.; Krowarsch, D.; Jelen, F.; Otlewski, J. A Conjugate Based on Anti-HER2 Diaffibody and Auristatin E Targets HER2-Positive Cancer Cells. Int. J. Mol. Sci. 2017, 18, 401. https://doi.org/10.3390/ijms18020401
Serwotka-Suszczak AM, Sochaj-Gregorczyk AM, Pieczykolan J, Krowarsch D, Jelen F, Otlewski J. A Conjugate Based on Anti-HER2 Diaffibody and Auristatin E Targets HER2-Positive Cancer Cells. International Journal of Molecular Sciences. 2017; 18(2):401. https://doi.org/10.3390/ijms18020401
Chicago/Turabian StyleSerwotka-Suszczak, Anna M., Alicja M. Sochaj-Gregorczyk, Jerzy Pieczykolan, Daniel Krowarsch, Filip Jelen, and Jacek Otlewski. 2017. "A Conjugate Based on Anti-HER2 Diaffibody and Auristatin E Targets HER2-Positive Cancer Cells" International Journal of Molecular Sciences 18, no. 2: 401. https://doi.org/10.3390/ijms18020401