Response of Gene Expression and Alternative Splicing to Distinct Growth Environments in Tomato
Abstract
:1. Introduction
2. Results
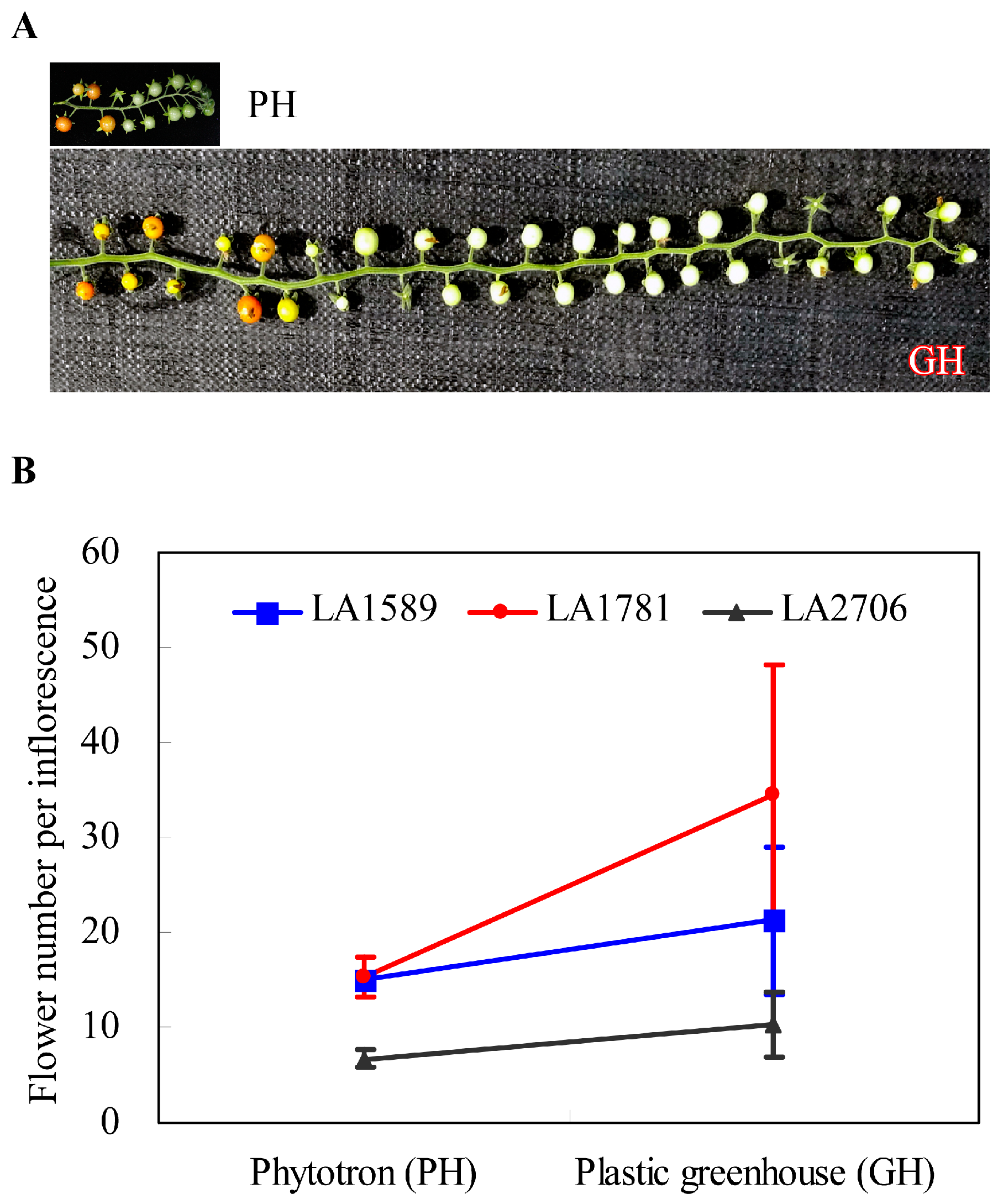
2.1. Characterization of Alternative Splicing Events in Tomato Seedlings Growing in Two Distinct Environments
2.2. Transcriptional Response to Environmental Changes
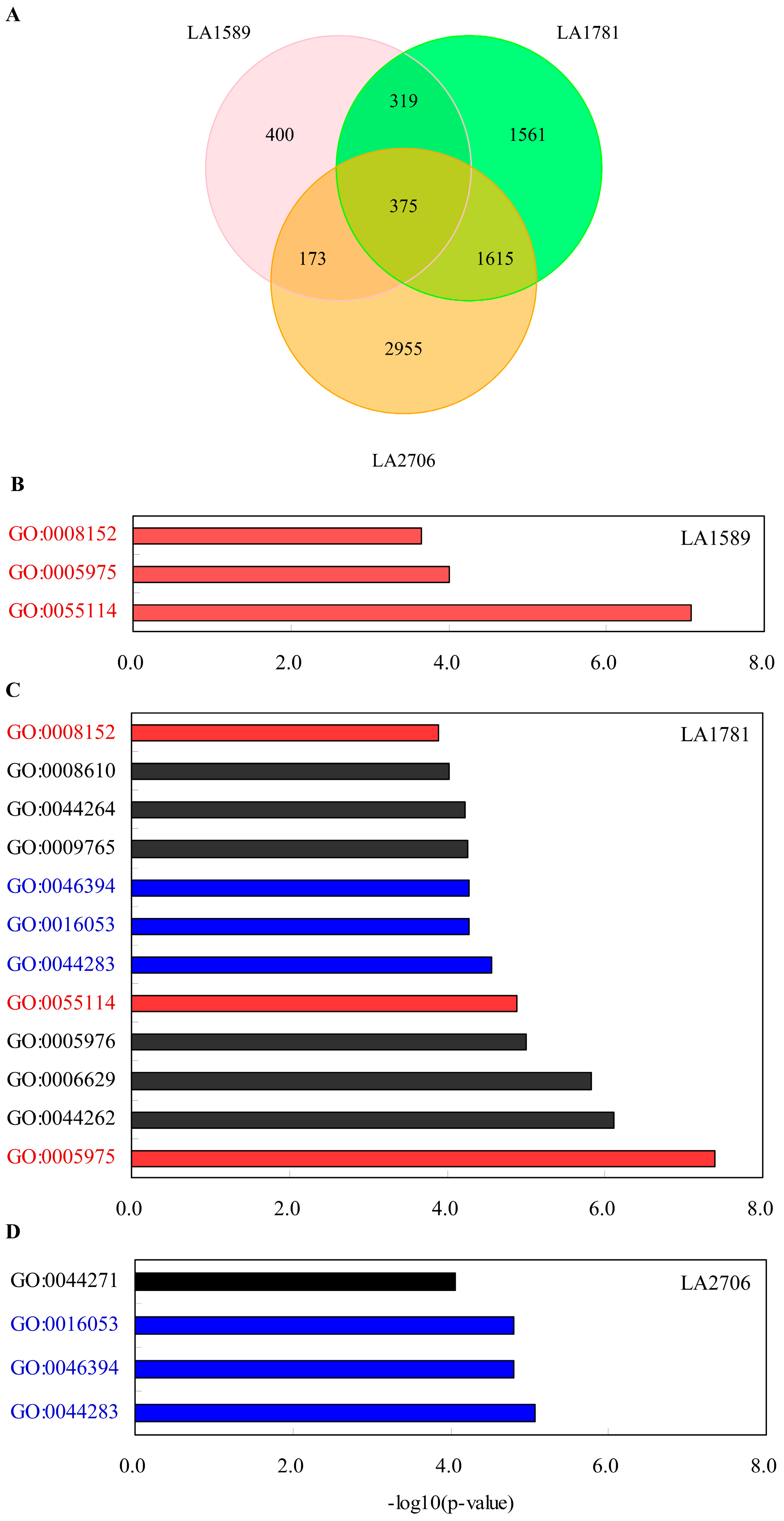
2.3. Identification of Environment-Regulated Genes in Tomato
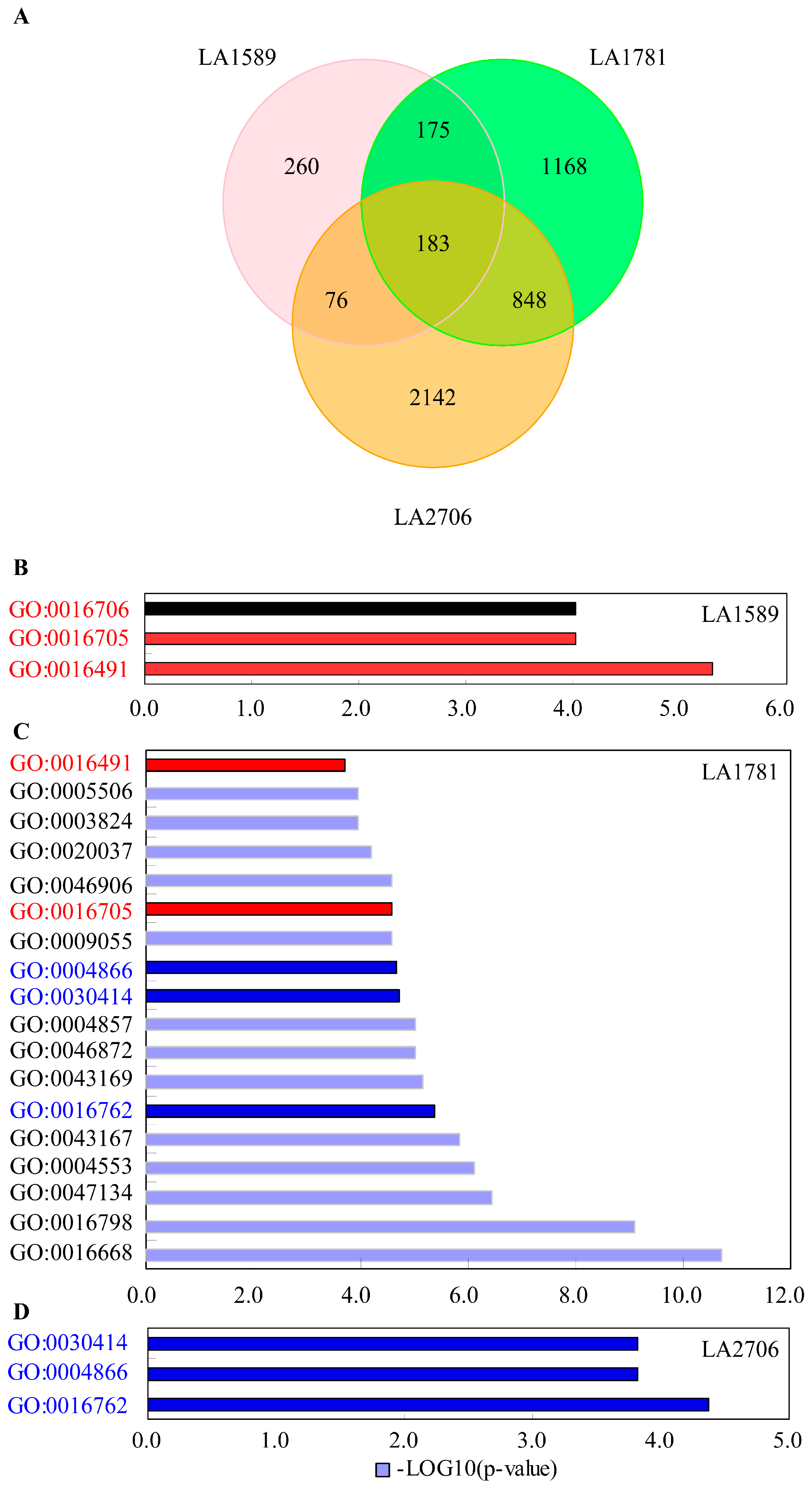
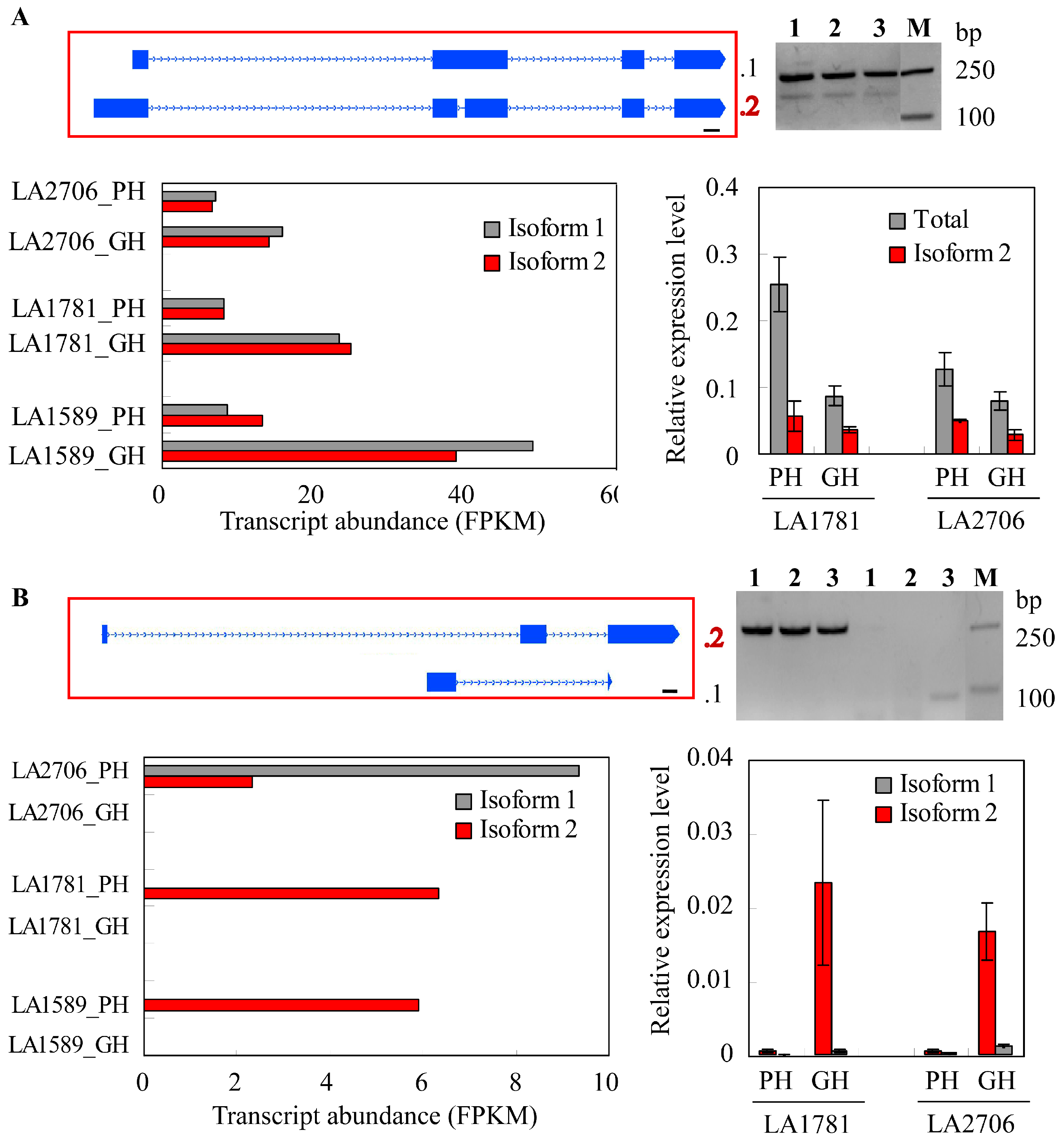
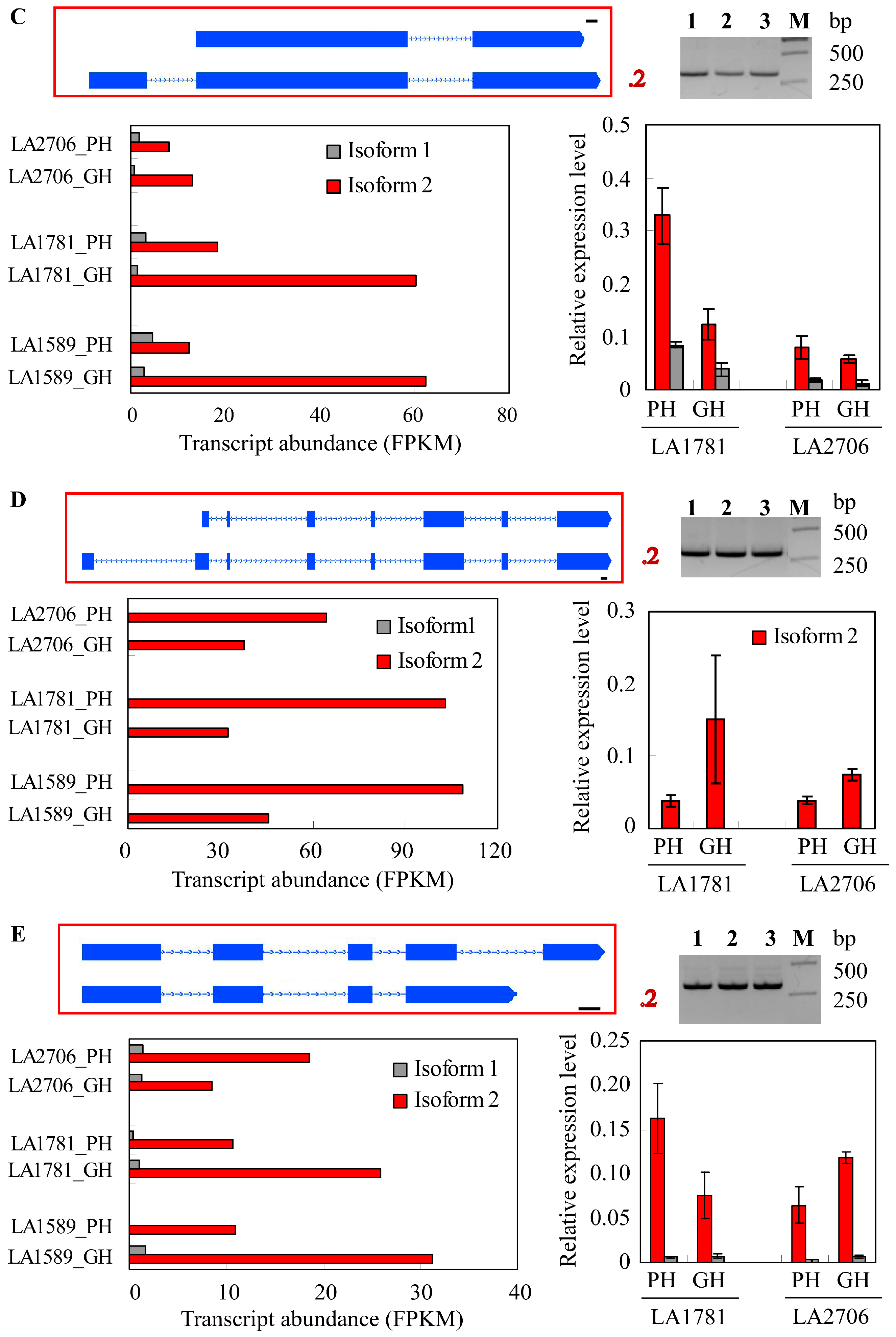
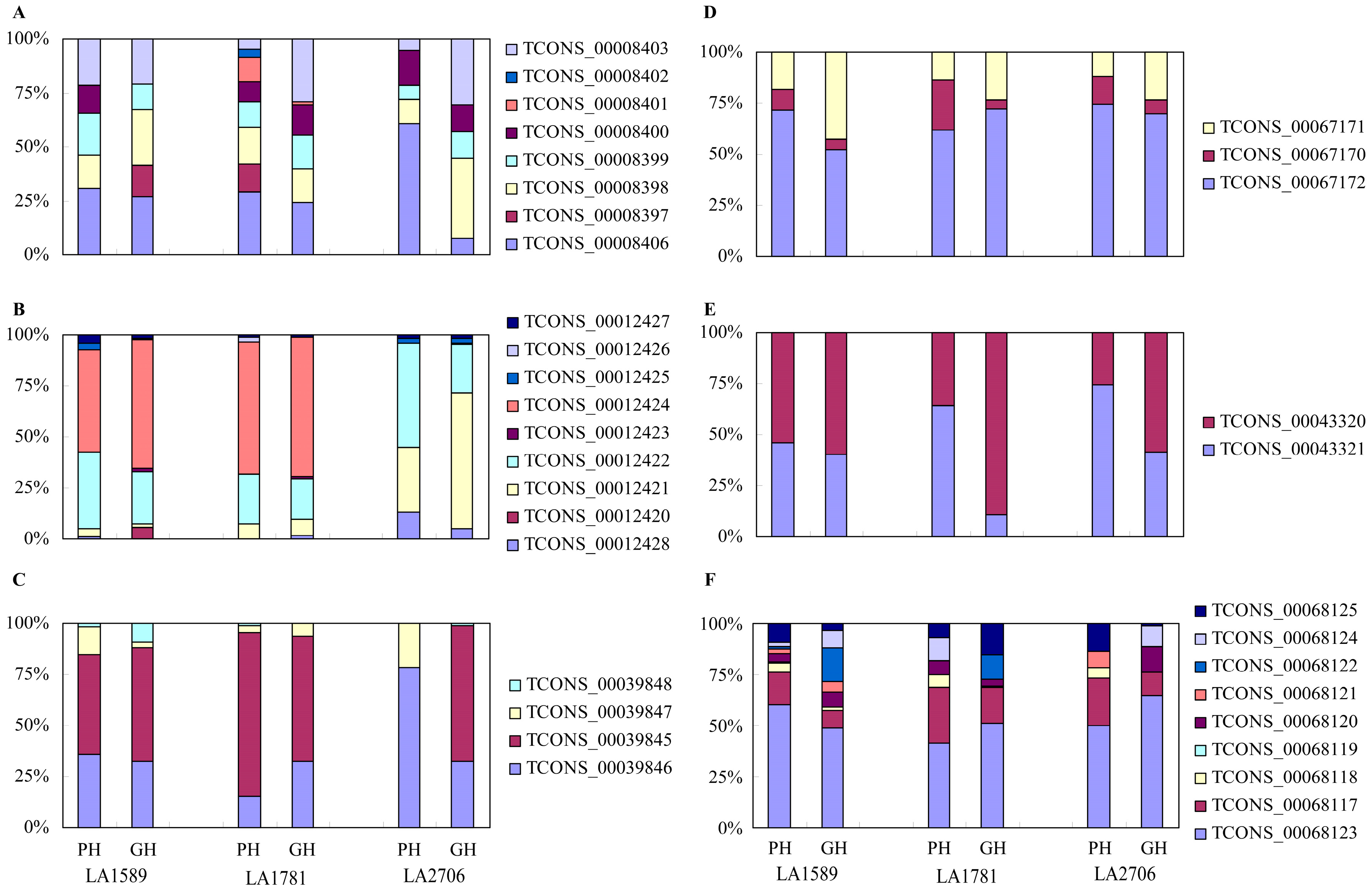
2.4. Response of Alternative Splicing to Environments
3. Discussion
3.1. Genotype-Dependent AS Regulation by Distinct Environments
3.2. Transcriptome Dynamics in Response to Environmental Fluctuations
4. Materials and Methods
4.1. Plant Growth Conditions
4.2. RNA Sequencing, Read Mapping and Transcript Assembly
4.3. Identification of AS Events
4.4. Gene Ontology Enrichment Analysis of Differentially Expressed Genes and Isoforms
4.5. Validation of Isoform Expression by RT-PCR and Quantitative RT-PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sultan, S.E. Phenotypic plasticity for plant development, function and life history. Trends Plant Sci. 2000, 5, 537–542. [Google Scholar] [CrossRef]
- Brosche, M.; Vinocur, B.; Alatalo, E.R.; Lamminmaki, A.; Teichmann, T.; Ottow, E.A.; Djilianov, D.; Afif, D.; Bogeat-Triboulot, M.B.; Altman, A.; et al. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biol. 2005, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Wildhagen, H.; Junker, L.V.; Ensminger, I. Transcriptome responses to temperature, water availability and photoperiod are conserved among mature trees of two divergent Douglas-fir provenances from a coastal and an interior habitat. BMC Genom. 2016, 17, 682. [Google Scholar] [CrossRef] [PubMed]
- Holliday, J.A.; Ralph, S.G.; White, R.; Bohlmann, J.; Aitken, S.N. Global monitoring of autumn gene expression within and among phenotypically divergent populations of Sitka spruce (Picea sitchensis). New Phytol. 2008, 178, 103–122. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.L.; Rosas, U.; Banta, J.; Bhambhra, N.; Purugganan, M.D. Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet. 2012, 8, e1002662. [Google Scholar] [CrossRef] [PubMed]
- Travers, S.E.; Tang, Z.; Caragea, D.; Garrett, K.A.; Hulbert, S.H.; Leach, J.E.; Bai, J.; Saleh, A.; Knapp, A.K.; Fay, P.A.; et al. Variation in gene expression of Andropogon gerardii in response to altered environmental conditions associated with climate change. J. Ecol. 2010, 98, 374–383. [Google Scholar] [CrossRef]
- Dal Santo, S.; Tornielli, G.B.; Zenoni, S.; Fasoli, M.; Farina, L.; Anesi, A.; Guzzo, F.; Delledonne, M.; Pezzotti, M. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013, 14, r54. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.J.; Sato, Y.; Mihara, M.; Antonio, B.A.; Motoyama, R.; Itoh, H.; Nagamura, Y.; Izawa, T. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell 2012, 151, 1358–1369. [Google Scholar] [CrossRef] [PubMed]
- Plessis, A.; Hafemeister, C.; Wilkins, O.; Gonzaga, Z.J.; Meyer, R.S.; Pires, I.; Muller, C.; Septiningsih, E.M.; Bonneau, R.; Purugganan, M. Multiple abiotic stimuli are integrated in the regulation of rice gene expression under field conditions. eLife 2015, 4, e08411. [Google Scholar] [CrossRef] [PubMed]
- Lei, B.; Lu, K.; Ding, F.; Zhang, K.; Chen, Y.; Zhao, H.; Zhang, L.; Ren, Z.; Qu, C.; Guo, W.; et al. RNA sequencing analysis reveals transcriptomic variations in tobacco (Nicotiana tabacum) leaves affected by climate, soil, and tillage factors. Int. J. Mol. Sci. 2014, 15, 6137–6160. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the alternative splicing landscape in plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Tian, C.; Yu, Y.; Jiao, Y. Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol. Plant 2016, 9, 749–752. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Crespi, M.; Bennett, M.J. One gene, many proteins: Mapping cell-specific alternative splicing in plants. Dev. Cell 2016, 39, 383–385. [Google Scholar] [CrossRef] [PubMed]
- De Klerk, E.; Hoen, P.A. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015, 31, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Syed, N.H.; Kalyna, M.; Marquez, Y.; Barta, A.; Brown, J.W. Alternative splicing in plants—Coming of age. Trends Plant Sci. 2012, 17, 616–623. [Google Scholar] [CrossRef] [PubMed]
- English, A.C.; Patel, K.S.; Loraine, A.E. Prevalence of alternative splicing choices in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 102. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Lu, G.; Fan, D.; Zhu, C.; Li, W.; Zhao, Q.; Feng, Q.; Zhao, Y.; Guo, Y.; Huang, X.; et al. Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res. 2010, 20, 1238–1249. [Google Scholar] [CrossRef] [PubMed]
- Marquez, Y.; Brown, J.W.; Simpson, C.; Barta, A.; Kalyna, M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012, 22, 1184–1195. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.; Peng, D.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xiao, H. Identification of alternative splicing events by RNA sequencing in early growth tomato fruits. BMC Genom. 2015, 16, 948. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-wide analysis of alternative splicing in Zea mays: Landscape and genetic regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef] [PubMed]
- Chamala, S.; Feng, G.; Chavarro, C.; Barbazuk, W.B. Genome-wide identification of evolutionarily conserved alternative splicing events in flowering plants. Front. Bioeng. Biotechnol. 2015, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Alshareef, S.; Butt, H.; Lozano-Juste, J.; Li, L.; Galal, A.A.; Moustafa, A.; Momin, A.A.; Tashkandi, M.; Richardson, D.N.; et al. Pre-mRNA splicing repression triggers abiotic stress signaling in plants. Plant J. 2017, 89, 291–309. [Google Scholar] [CrossRef] [PubMed]
- Capovilla, G.; Pajoro, A.; Immink, R.G.; Schmid, M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015, 27, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.W.; Simpson, C.G.; Marquez, Y.; Gadd, G.M.; Barta, A.; Kalyna, M. Lost in translation: Pitfalls in deciphering plant alternative splicing transcripts. Plant Cell 2015, 27, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Tang, F.; Zhu, H. Alternative splicing in plant immunity. Int. J. Mol. Sci. 2014, 15, 10424–10445. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–5366. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, L.; Drewe-Boss, P.; Wiessner, T.; Wagner, G.; Geue, S.; Lee, H.C.; Obermuller, D.M.; Kahles, A.; Behr, J.; Sinz, F.H.; et al. Alternative splicing substantially diversifies the transcriptome during early photomorphogenesis and correlates with the energy availability in Arabidopsis. Plant Cell 2016, 28, 2715–2734. [Google Scholar] [CrossRef] [PubMed]
- Ner-Gaon, H.; Halachmi, R.; Savaldi-Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004, 39, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Cumbie, J.S.; Dharmawardhana, P.; Jaiswal, P.; Chang, J.H.; Palusa, S.G.; Reddy, A.S.; Megraw, M.; Mockler, T.C. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant 2015, 8, 207–227. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.P.; Su, Y.S.; Chen, H.C.; Chen, Y.R.; Wu, C.C.; Lin, W.D.; Tu, S.L. Genome-wide analysis of light-regulated alternative splicing mediated by photoreceptors in Physcomitrella patens. Genome Biol. 2014, 15, R10. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Lin, W.D.; Tu, S.L. Genome-wide analysis of heat-sensitive alternative splicing in Physcomitrella patens. Plant Physiol. 2014, 165, 826–840. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Cui, P.; Wang, Z.; Zhang, S.; Ali, S.; Xiong, L. Genome-wide analysis of alternative splicing of pre-mRNA under salt stress in Arabidopsis. BMC Genom. 2014, 15, 431. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.; Priest, H.D.; Megraw, M.; Mockler, T.C. Alternative splicing in plants: Directing traffic at the crossroads of adaptation and environmental stress. Curr. Opin. Plant Biol. 2015, 24, 125–135. [Google Scholar] [CrossRef] [PubMed]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.; Nimmo, H.G. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Mandadi, K.K.; Scholthof, K.B. Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 2015, 27, 71–85. [Google Scholar] [CrossRef] [PubMed]
- Shikata, H.; Hanada, K.; Ushijima, T.; Nakashima, M.; Suzuki, Y.; Matsushita, T. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 18781–18786. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Xu, Q.; Wang, X. Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 2014, 65, 1973–1980. [Google Scholar] [CrossRef] [PubMed]
- Palusa, S.G.; Ali, G.S.; Reddy, A.S. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: Regulation by hormones and stresses. Plant J. 2007, 49, 1091–1107. [Google Scholar] [CrossRef] [PubMed]
- Dzakovich, M.P.; Ferruzzi, M.G.; Mitchell, C.A. Manipulating sensory and phytochemical profiles of greenhouse tomatoes using environmentally relevant doses of ultraviolet radiation. J. Agric. Food Chem. 2016, 64, 6801–6808. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Sun, J.; Shu, S.; Lu, X.; Tian, J.; Wang, J. Analysis of general situation, characteristics, existing problems and development trend of protected horticulture in China. China Veg. 2012, 18, 1–14. [Google Scholar]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Lou, Y.; Sun, H.; Li, L.; Wang, L.; Dong, L.; Gao, Z. Transcriptome and comparative gene expression analysis of Phyllostachys edulis in response to high light. BMC Plant Biol. 2016, 16, 34. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Okamoto, H.; Iwasaki, Y.; Asahi, T. A deficiency of coproporphyrinogen III oxidase causes lesion formation in Arabidopsis. Plant J. 2001, 27, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Naested, H.; Holm, A.; Jenkins, T.; Nielsen, H.B.; Harris, C.A.; Beale, M.H.; Andersen, M.; Mant, A.; Scheller, H.; Camara, B.; et al. Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J. Cell Sci. 2004, 117, 4807–4818. [Google Scholar] [CrossRef] [PubMed]
- Bolle, C.; Koncz, C.; Chua, N.H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 2000, 14, 1269–1278. [Google Scholar] [PubMed]
- Gonzalez Besteiro, M.A.; Ulm, R. ATR and MKP1 play distinct roles in response to UV-B stress in Arabidopsis. Plant J. 2013, 73, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Torres-Galea, P.; Hirtreiter, B.; Bolle, C. Two GRAS proteins, SCARECROW-LIKE 21 and PHYTOCHROME A SIGNAL TRANSDUCTION1, function cooperatively in phytochrome A signal transduction. Plant Physiol. 2013, 161, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Ulm, R.; Revenkova, E.; di Sansebastiano, G.P.; Bechtold, N.; Paszkowski, J. Mitogen-activated protein kinase phosphatase is required for genotoxic stress relief in Arabidopsis. Genes Dev. 2001, 15, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lin, C.; Gu, L. Light regulation of alternative pre-mRNA splicing in plants. Photochem. Photobiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, R.F.; Szakonyi, D.; Simpson, C.G.; Barbosa, I.C.; Brown, J.W.; Baena-Gonzalez, E.; Duque, P. The Arabidopsis SR45 splicing factor, a negative regulator of sugar signaling, modulates SNF1-related protein kinase 1 stability. Plant Cell 2016, 28, 1910–1925. [Google Scholar] [CrossRef] [PubMed]
- Vitulo, N.; Forcato, C.; Carpinelli, E.C.; Telatin, A.; Campagna, D.; D’Angelo, M.; Zimbello, R.; Corso, M.; Vannozzi, A.; Bonghi, C.; et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014, 14, 99. [Google Scholar] [CrossRef] [PubMed]
- Cruz, T.M.; Carvalho, R.F.; Richardson, D.N.; Duque, P. Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 2014, 15, 17541–17564. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, M.J.; Park, C.M. Alternative splicing of transcription factors in plant responses to low temperature stress: Mechanisms and functions. Planta 2013, 237, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, C.; Cao, C.; Zhang, S.; Cai, H. Application research of microclimate change characteristic AD early warning service system in vegetable greenhouse. Acta Agric. Shanghai 2015, 31, 69–74. [Google Scholar]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [Green Version]
- Palusa, S.G.; Reddy, A.S. Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 2009, 185, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Drechsel, G.; Kahles, A.; Kesarwani, A.K.; Stauffer, E.; Behr, J.; Drewe, P.; Ratsch, G.; Wachter, A. Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 2013, 25, 3726–3742. [Google Scholar] [CrossRef] [PubMed]
- Kalyna, M.; Simpson, C.G.; Syed, N.H.; Lewandowska, D.; Marquez, Y.; Kusenda, B.; Marshall, J.; Fuller, J.; Cardle, L.; McNicol, J.; et al. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2012, 40, 2454–2469. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, T.; Douglass, S.; Gabunilas, J.; Pellegrini, M.; Chanfreau, G.F. Widespread use of non-productive alternative splice sites in Saccharomyces cerevisiae. PLoS Genet. 2014, 10, e1004249. [Google Scholar] [CrossRef] [PubMed]
- Lareau, L.F.; Brenner, S.E. Regulation of splicing factors by alternative splicing and NMD is conserved between kingdoms yet evolutionarily flexible. Mol. Biol. Evol. 2015, 32, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Sureshkumar, S.; Dent, C.; Seleznev, A.; Tasset, C.; Balasubramanian, S. Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2016, 2, 16055. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Campbell, T.G.; Stone, E.A.; Mackay, T.F.; Anholt, R.R. Phenotypic plasticity of the Drosophila transcriptome. PLoS Genet. 2012, 8, e1002593. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007, 35, 297–299. [Google Scholar] [CrossRef] [PubMed]

| ITAG2.4 ID | Fold Changes (log2) | Description | ||
|---|---|---|---|---|
| LA1589 | LA1781 | LA2706 | ||
| Solyc11g068430 | −2.95 | −2.74 | −1.03 | Ferredoxin |
| Solyc06g069730 | −1.30 | −3.34 | −1.28 | Chlorophyll a–b binding protein 4, chloroplastic |
| Solyc05g056050 | −1.22 | −2.34 | −1.43 | Chlorophyll a–b binding protein |
| Solyc08g006930 | −1.16 | −1.52 | −1.09 | Photosystem I reaction center subunit X psaK |
| Solyc12g006140 | −1.14 | −3.16 | −1.48 | Chlorophyll a–b binding protein 37 |
| Solyc02g077240 | −5.43 | −3.96 | −2.01 | Pyruvate decarboxylase |
| Solyc10g076510 | −3.88 | −2.02 | −1.04 | Pyruvate decarboxylase |
| Solyc06g048410 | −1.76 | −2.15 | −1.05 | Superoxide dismutase |
| Solyc03g120990 | −1.24 | −2.50 | −2.12 | Malic enzyme; Malic enzyme, NAD-binding |
| Solyc01g095140 | −2.80 | −2.34 | −1.04 | Late embryogenesis abundant protein; response to light |
| Solyc10g047530 | −1.29 | −2.80 | −1.28 | Phototropic-responsive NPH3 family protein |
| Solyc01g109700 | 1.00 | 1.31 | 1.01 | Transcription factor; Helix-loop-helix DNA-binding |
| Solyc06g007180 | 1.78 | 4.10 | 3.72 | Asparagine synthase (Glutamine-hydrolyzing) |
| Solyc05g008240 | infinite * | infinite | infinite | myb3 like; similar to R2R3-myb anthocyanin repressor |
| Functional Category | S. Pimpinellifolium | S. Lycopersicum | |
|---|---|---|---|
| LA1589 | LA1781 | LA2706 | |
| DNA modification | 2 (4.5) * | 2 (2.8) | 3 (2.3) |
| RNA metabolism | 3 (6.8) | 8 (11.3) | 14 (10.9) |
| Protein biosynthesis and modification | 10 (22.7) | 11 (15.5) | 14 (10.9) |
| Response to stresses | 6 (13.6) | 7 (9.9) | 8 (6.2) |
| Growth and development | 5 (11.4) | 5 (7.0) | 18 (14.0) |
| Photosynthesis and light signaling | - | 4 (5.6) | 5 (3.9) |
| miscellaneous | 3 (6.8) | 8 (11.3) | 14 (10.9) |
| Unknown | 44 (34.1) | 26 (36.6) | 53 (41.1) |
| Total | 44 (100) | 71 (100) | 129 (100) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Weng, L.; Li, M.; Xiao, H. Response of Gene Expression and Alternative Splicing to Distinct Growth Environments in Tomato. Int. J. Mol. Sci. 2017, 18, 475. https://doi.org/10.3390/ijms18030475
Wang G, Weng L, Li M, Xiao H. Response of Gene Expression and Alternative Splicing to Distinct Growth Environments in Tomato. International Journal of Molecular Sciences. 2017; 18(3):475. https://doi.org/10.3390/ijms18030475
Chicago/Turabian StyleWang, Guixiang, Lin Weng, Meng Li, and Han Xiao. 2017. "Response of Gene Expression and Alternative Splicing to Distinct Growth Environments in Tomato" International Journal of Molecular Sciences 18, no. 3: 475. https://doi.org/10.3390/ijms18030475







