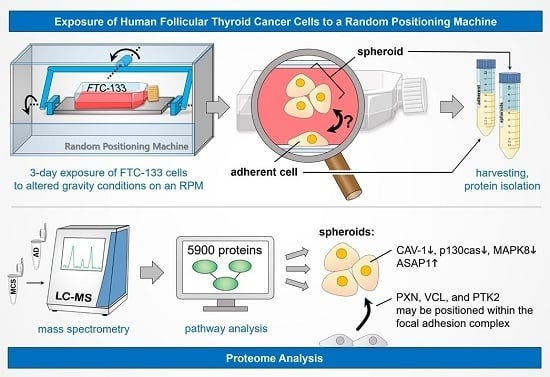
Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine
Abstract
:1. Introduction
2. Results and Discussion
2.1. Spheroid Formation
2.2. Quantitative Overview on Proteins Detected by Mass Spectrometry
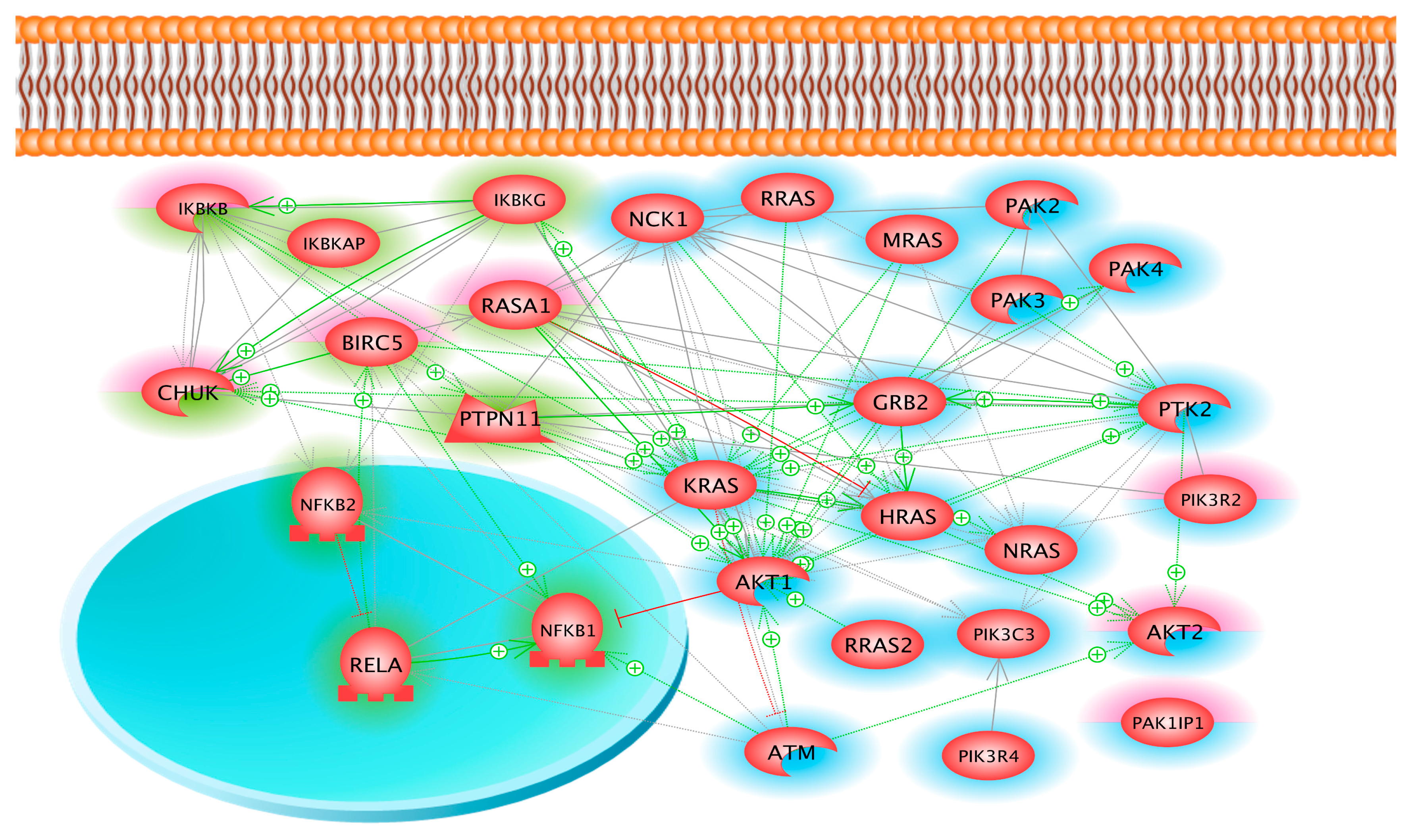
2.3. Coverage of Different Canonical Pathways by the Proteins Detected in Different Cell Samples
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Random Positioning Machine
4.3. Cell Harvest
4.4. Mass Spectometry
4.5. Pathway Analysis
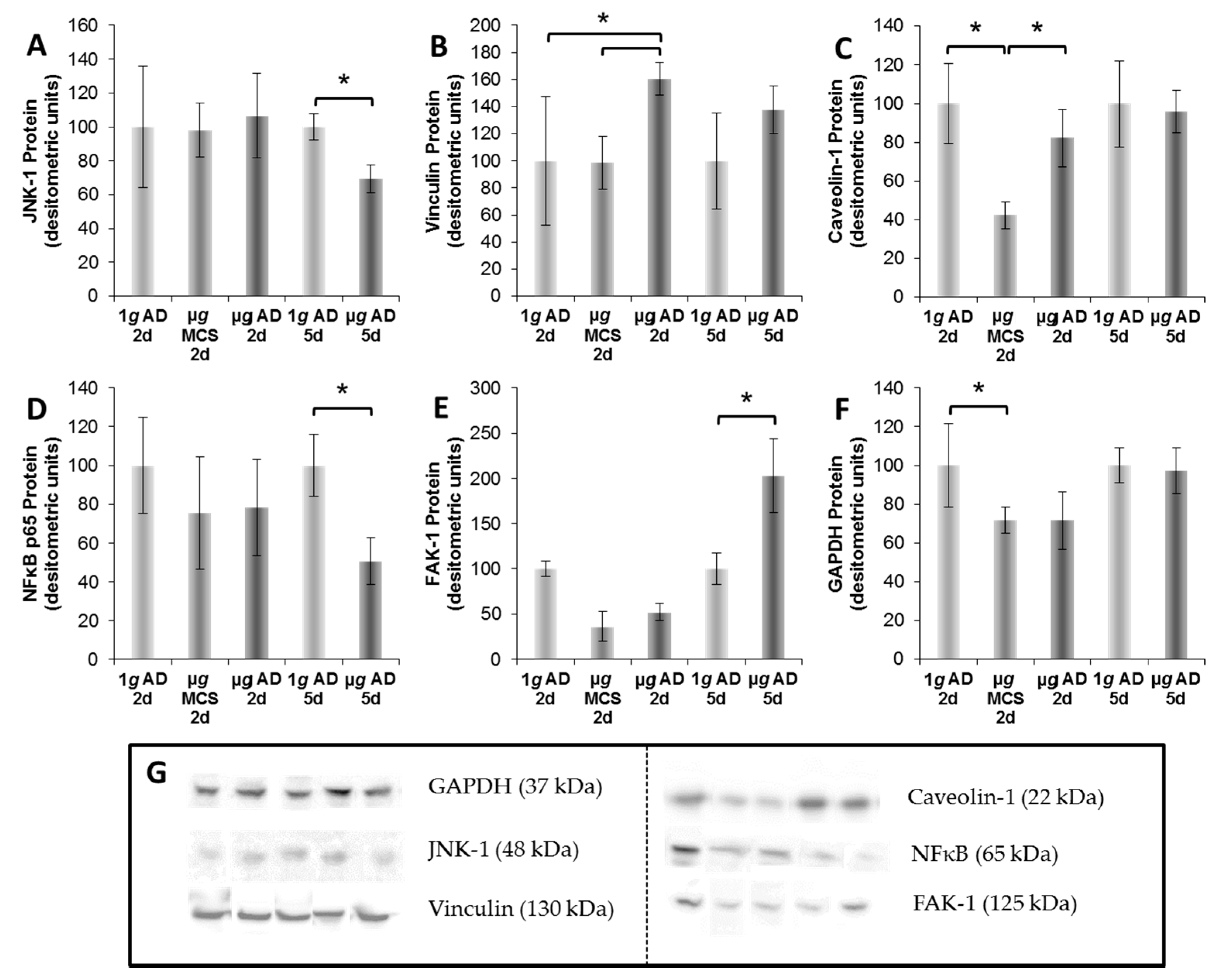
4.6. Western Blot
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AD | Adherent cells |
| AKT1 | RAC-alpha serine/threonine-protein kinase 1 |
| ARF6 | ADP-ribosylation factor 6 |
| ASAP1 | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1 |
| BIRC5 | Baculoviral inhibitor of apoptosis repeat-containing 5; Survivin |
| CAV-1 | Caveolin 1 |
| CHUK | Conserved helix-loop-helix ubiquitous kinase; Inhibitor of nuclear factor kappa-B kinase subunit alpha (IKK-α) |
| GAPDH | Glycerinaldehyd-3-phosphat-Dehydrogenase |
| HRP | Horseradish peroxidase |
| IKBKB | Inhibitor of nuclear factor kappa-B kinase subunit beta |
| JNK1 | c-Jun N-terminal kinase 1 or Mitogen-activated protein kinase 8 |
| LfQ | Label-free Quantitation |
| MAPK8 | Mitogen-activated protein kinase 8 |
| MCS | Multicellular tumor spheroids |
| MS | Mass Spectrometry |
| NFκB | Nuclear factor kappa B |
| PAK1IP1 | P21-activated protein kinase-interacting protein 1 |
| PIK3R2 | Phosphoinositide-3-kinase regulatory subunit 2 |
| PTK2 | Protein tyrosine kinase 2, focal adhesion kinase 1 (FAK1) |
| PXN | Paxillin |
| Ras1 | Ras-like protein 1 |
| r-µg | Real microgravity |
| RPM | Random positioning machine |
| s-µg | Simulated microgravity |
| VCL | Vinculin |
| VEGF | Vascular endothelial growth factor |
References
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Chatzinikolaidou, M. Cell spheroids: The new frontiers in in vitro models for cancer drug validation. Drug Discov. Today 2016, 21, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Wehland, M.; Pietsch, J.; Aleshcheva, G.; Wise, P.; van Loon, J.; Ulbrich, C.; Magnusson, N.E.; Infanger, M.; Bauer, J. Growing tissues in real and simulated microgravity: New methods for tissue engineering. Tissue Eng. Part B Rev. 2014, 20, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.L.; Souza, G.R. Using space-based investigations to inform cancer research on Earth. Nat. Rev. Cancer 2013, 13, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Pietsch, J.; Wehland, M.; Schulz, H.; Saar, K.; Hubner, N.; Bauer, J.; Braun, M.; Schwarzwalder, A.; Segerer, J.; et al. Differential gene expression profile and altered cytokine secretion of thyroid cancer cells in space. FASEB J. 2014, 28, 813–835. [Google Scholar] [CrossRef] [PubMed]
- Warnke, E.; Pietsch, J.; Wehland, M.; Bauer, J.; Infanger, M.; Gorog, M.; Hemmersbach, R.; Braun, M.; Ma, X.; Sahana, J.; et al. Spheroid formation of human thyroid cancer cells under simulated microgravity: A possible role of CTGF and CAV1. Cell Commun. Signal. 2014, 12, 32. [Google Scholar] [CrossRef] [PubMed]
- Riwaldt, S.; Pietsch, J.; Sickmann, A.; Bauer, J.; Braun, M.; Segerer, J.; Schwarzwalder, A.; Aleshcheva, G.; Corydon, T.J.; Infanger, M.; et al. Identification of proteins involved in inhibition of spheroid formation under microgravity. Proteomics 2015, 15, 2945–2952. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.; Warnke, E.; Wehland, M.; Aleshcheva, G.; Magnusson, N.E.; Hemmersbach, R.; Corydon, T.J.; Bauer, J.; Infanger, M.; Grimm, D. Mechanisms of three-dimensional growth of thyroid cells during long-term simulated microgravity. Sci. Rep. 2015, 5, 16691. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Wehland, M.; Pietsch, J.; Sickmann, A.; Weber, G.; Grimm, D. Annotated Gene and Proteome Data Support Recognition of Interconnections Between the Results of Different Experiments in Space Research. Microgravity Sci. Technol. 2016, 28, 357–365. [Google Scholar] [CrossRef]
- Riwaldt, S.; Bauer, J.; Pietsch, J.; Braun, M.; Segerer, J.; Schwarzwalder, A.; Corydon, T.J.; Infanger, M.; Grimm, D. The Importance of Caveolin-1 as Key-Regulator of Three-Dimensional Growth in Thyroid Cancer Cells Cultured under Real and Simulated Microgravity Conditions. Int. J. Mol. Sci. 2015, 16, 28296–28310. [Google Scholar] [CrossRef] [PubMed]
- Grosse, J.; Wehland, M.; Pietsch, J.; Schulz, H.; Saar, K.; Hubner, N.; Eilles, C.; Bauer, J.; Abou-El-Ardat, K.; Baatout, S.; et al. Gravity-sensitive signaling drives 3-dimensional formation of multicellular thyroid cancer spheroids. FASEB J. 2012, 26, 5124–5140. [Google Scholar] [CrossRef] [PubMed]
- Kossmehl, P.; Shakibaei, M.; Cogoli, A.; Infanger, M.; Curcio, F.; Schonberger, J.; Eilles, C.; Bauer, J.; Pickenhahn, H.; Schulze-Tanzil, G.; et al. Weightlessness induced apoptosis in normal thyroid cells and papillary thyroid carcinoma cells via extrinsic and intrinsic pathways. Endocrinology 2003, 144, 4172–4179. [Google Scholar] [CrossRef] [PubMed]
- Mann, M.; Kulak, N.A.; Nagaraj, N.; Cox, J. The coming age of complete, accurate, and ubiquitous proteomes. Mol. Cell 2013, 49, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, J.; Kussian, R.; Sickmann, A.; Bauer, J.; Weber, G.; Nissum, M.; Westphal, K.; Egli, M.; Grosse, J.; Schonberger, J.; et al. Application of free-flow IEF to identify protein candidates changing under microgravity conditions. Proteomics 2010, 10, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, J.; Bauer, J.; Weber, G.; Nissum, M.; Westphal, K.; Egli, M.; Grosse, J.; Schönberger, J.; Eilles, C.; Infanger, M.; et al. Proteome Analysis of Thyroid Cancer Cells After Long-Term Exposure to a Random Positioning Machine. Microgravity Sci. Technol. 2011, 23, 381–390. [Google Scholar] [CrossRef]
- Obermaier, C.; Jankowski, V.; Schmutzler, C.; Bauer, J.; Wildgruber, R.; Infanger, M.; Kohrle, J.; Krause, E.; Weber, G.; Grimm, D. Free-flow isoelectric focusing of proteins remaining in cell fragments following sonication of thyroid carcinoma cells. Electrophoresis 2005, 26, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Svejgaard, B.; Wehland, M.; Ma, X.; Kopp, S.; Sahana, J.; Warnke, E.; Aleshcheva, G.; Hemmersbach, R.; Hauslage, J.; Grosse, J.; et al. Common Effects on Cancer Cells Exerted by a Random Positioning Machine and a 2D Clinostat. PLoS ONE 2015, 10, e0135157. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, J.; Sickmann, A.; Weber, G.; Bauer, J.; Egli, M.; Wildgruber, R.; Infanger, M.; Grimm, D. A proteomic approach to analysing spheroid formation of two human thyroid cell lines cultured on a random positioning machine. Proteomics 2011, 11, 2095–2104. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, P.A.; Andrade, J.; Miura, K.; Brown, M.T.; Long, Y.Q.; Stauffer, S.; Roller, P.; Cooper, J.A. The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 2000, 97, 4011–4016. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yerushalmi, G.M.; Grigera, P.R.; Parsons, J.T. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 2005, 280, 8884–8892. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Wada, I.; Miura, K.; Okawa, K.; Kadoya, T.; Kato, T.; Nishihara, H.; Maeda, M.; Tanaka, S.; Nagashima, K.; et al. CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 2003, 278, 6456–6460. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.E.; Glenney, J.R., Jr.; Burridge, K. Paxillin: A new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990, 111, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Loijens, J.C.; Martin, K.H.; Karginov, A.V.; Parsons, J.T. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell 2002, 13, 2147–2156. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, P.A.; Hirsch, D.S. Arf GAPs: Multifunctional proteins that regulate membrane traffic and actin remodelling. Cell Signal. 2004, 16, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Sabe, H.; Onodera, Y.; Mazaki, Y.; Hashimoto, S. ArfGAP family proteins in cell adhesion, migration and tumor invasion. Curr. Opin. Cell Biol. 2006, 18, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Deakin, N.O.; Turner, C.E. Paxillin comes of age. J. Cell Sci. 2008, 121, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Zaidel-Bar, R.; Kam, Z.; Geiger, B. Polarized downregulation of the paxillin-p130CAS-Rac1 pathway induced by shear flow. J. Cell Sci. 2005, 118, 3997–4007. [Google Scholar] [CrossRef] [PubMed]
- Kumbrink, J.; Soni, S.; Laumbacher, B.; Loesch, B.; Kirsch, K.H. Identification of Novel Crk-associated Substrate (p130Cas) Variants with Functionally Distinct Focal Adhesion Kinase Binding Activities. J. Biol. Chem. 2015, 290, 12247–12255. [Google Scholar] [CrossRef] [PubMed]
- Harte, M.T.; Hildebrand, J.D.; Burnham, M.R.; Bouton, A.H.; Parsons, J.T. p130Cas, a substrate associated with v-Src and v-Crk, localizes to focal adhesions and binds to focal adhesion kinase. J. Biol. Chem. 1996, 271, 13649–13655. [Google Scholar] [PubMed]
- Goldmann, W.H. Vinculin-p130Cas interaction is critical for focal adhesion dynamics and mechano-transduction. Cell Biol. Int. 2014, 38, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Goldmann, W.H. Role of vinculin in cellular mechanotransduction. Cell Biol. Int. 2016, 40, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kim, M.; Hu, Y.L.; Jalali, S.; Schlaepfer, D.D.; Hunter, T.; Chien, S.; Shyy, J.Y. Fluid shear stress activation of focal adhesion kinase. Linking to mitogen-activated protein kinases. J. Biol. Chem. 1997, 272, 30455–30462. [Google Scholar] [CrossRef] [PubMed]
- Bourmoum, M.; Charles, R.; Claing, A. The GTPase ARF6 Controls ROS Production to Mediate Angiotensin II-Induced Vascular Smooth Muscle Cell Proliferation. PLoS ONE 2016, 11, e0148097. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Levy, D.; Ruiz, J.L.; Brocardo, G.A.; Ferreira, K.A.; Costa, R.O.; Queiroz, R.G.; Maria, D.A.; Neto, A.E.; Chamone, D.A.; et al. Azidothymidine is effective against human multiple myeloma: A new use for an old drug? Anticancer Agents Med. Chem. 2013, 13, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Kopp, S.; Slumstrup, L.; Corydon, T.J.; Sahana, J.; Aleshcheva, G.; Islam, T.; Magnusson, N.E.; Wehland, M.; Bauer, J.; Infanger, M.; et al. Identifications of novel mechanisms in breast cancer cells involving duct-like multicellular spheroid formation after exposure to the Random Positioning Machine. Sci. Rep. 2016, 6, 26887. [Google Scholar] [CrossRef] [PubMed]
- Grimm, D.; Infanger, M.; Westphal, K.; Ulbrich, C.; Pietsch, J.; Kossmehl, P.; Vadrucci, S.; Baatout, S.; Flick, B.; Paul, M.; et al. A delayed type of three-dimensional growth of human endothelial cells under simulated weightlessness. Tissue Eng. Part A 2009, 15, 2267–2275. [Google Scholar] [CrossRef] [PubMed]
- Riwaldt, S.; Bauer, J.; Wehland, M.; Slumstrup, L.; Kopp, S.; Warnke, E.; Dittrich, A.; Magnusson, N.E.; Pietsch, J.; Corydon, T.J.; et al. Pathways Regulating Spheroid Formation of Human Follicular Thyroid Cancer Cells under Simulated Microgravity Conditions: A Genetic Approach. Int. J. Mol. Sci. 2016, 17, 528. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, A.; Sakakura, J.; Yagi, R.; Mazaki, Y.; Schaefer, E.; Yano, H.; Sabe, H. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 2002, 159, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.A.; Deakin, N.O.; Turner, C.E. Emerging role of paxillin-PKL in regulation of cell adhesion, polarity and migration. Cell Adh. Migr. 2010, 4, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Gigoux, V.; L’Hoste, S.; Raynaud, F.; Camonis, J.; Garbay, C. Identification of Aurora kinases as RasGAP Src homology 3 domain-binding proteins. J. Biol. Chem. 2002, 277, 23742–23746. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; An, J.; Shan, L.; Jiang, Q.; Li, F.; Ci, Y.; Wu, P.; Duan, J.; Hui, K.; Yang, Y.; et al. Survivin-2B promotes autophagy by accumulating IKK α in the nucleus of selenite-treated NB4 cells. Cell Death Dis. 2014, 5, e1071. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ghosh, S. Differential phosphorylation of the signal-responsive domain of IκBα and IκBβ by IκB kinases. J. Biol. Chem. 2003, 278, 31980–31987. [Google Scholar] [CrossRef] [PubMed]
- Corydon, T.J.; Kopp, S.; Wehland, M.; Braun, M.; Schutte, A.; Mayer, T.; Hulsing, T.; Oltmann, H.; Schmitz, B.; Hemmersbach, R.; et al. Alterations of the cytoskeleton in human cells in space proved by life-cell imaging. Sci. Rep. 2016, 6, 20043. [Google Scholar] [CrossRef] [PubMed]
- Goretzki, P.E.; Frilling, A.; Simon, D.; Roeher, H.D. Growth regulation of normal thyroids and thyroid tumors in man. Recent Results Cancer Res. 1990, 118, 48–63. [Google Scholar] [PubMed]
- Borst, A.G.; van Loon, J.J.W.A. Technology and Developments for the Random Positioning Machine, RPM. Microgravity Sci. Technol. 2008, 21, 287. [Google Scholar] [CrossRef]
- Warnke, E.; Kopp, S.; Wehland, M.; Hemmersbach, R.; Bauer, J.; Pietsch, J.; Infanger, M.; Grimm, D. Thyroid Cells Exposed to Simulated Microgravity Conditions—Comparison of the Fast Rotating Clinostat and the Random Positioning Machine. Microgravity Sci. Technol. 2016, 28, 247–260. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.; Kulak, N.A.; Cox, J.; Neuhauser, N.; Mayr, K.; Hoerning, O.; Vorm, O.; Mann, M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Analyzing Gels and Western Blots with ImageJ. Available online: http://lukemiller.org/index.php/2010/11/analyzing-gels-and-western-blots-with-image-j/ (accessed on 26 October 2016).

| Culture Condition | 1 | 2 and 3 | 4 | 5 and 6 | |
|---|---|---|---|---|---|
| Sample Number | #1 | #2 | #3 | #4 | #5 |
| Pre-incubation | 2 days | 2 days | 5 days | 5 days | |
| Following 3 days incubation under… | 1g | s-µg | 1g | s-µg | |
| Kind of growth at time of harvest | Adherent | Spheroid | Adherent | Adherent | Adherent |
| Number of proteins detected | 4419 | 4505 | 4544 | 4621 | 4961 |
| Cellular Compartment | Sample 1 | Sample 2 | Sample 3 | Sample 4 | Sample 5 |
|---|---|---|---|---|---|
| Secreted proteins | 19% | 19% | 19% | 20% | 21% |
| Membrane | 27% | 27% | 27% | 28% | 29% |
| Nucleus | 29% | 29% | 29% | 30% | 31% |
| Cytoplasm | 29% | 30% | 30% | 30% | 32% |
| Golgi apparatus | 31% | 31% | 32% | 34% | 35% |
| Cytoskeleton | 33% | 33% | 34% | 35% | 37% |
| Endoplasmic reticulum membrane | 35% | 36% | 37% | 36% | 39% |
| Endoplasmic reticulum | 36% | 36% | 37% | 36% | 39% |
| Perinuclear region of cytoplasm | 37% | 38% | 37% | 38% | 40% |
| Intracellular-membrane-bounded organelle | 37% | 37% | 38% | 39% | 40% |
| Endosome | 38% | 38% | 38% | 40% | 41% |
| Cytosol | 38% | 39% | 39% | 40% | 42% |
| Extracellular exosome | 41% | 43% | 42% | 44% | 45% |
| Nucleoplasm | 42% | 42% | 42% | 42% | 45% |
| Mitochondrion | 49% | 48% | 49% | 48% | 50% |
| Nucleolus | 50% | 50% | 50% | 49% | 50% |
| Nuclear speck | 51% | 50% | 50% | 50% | 57% |
| Mitochondrial inner membrane | 60% | 60% | 61% | 61% | 62% |
| Focal adhesion | 63% | 64% | 65% | 65% | 68% |
| Mitochondrial matrix | 65% | 62% | 63% | 65% | 63% |
| Spliceosomal complex | 68% | 69% | 66% | 66% | 71% |
| Ribonucleoprotein complex | 72% | 72% | 71% | 72% | 73% |
| Mitochondrial small ribosomal subunit | 93% | 93% | 93% | 93% | 93% |
| Large ribosomal subunit | 100% | 100% | 100% | 100% | 100% |
| Antibody | Dilution | Company | Molecular Weight | Catalog Number |
|---|---|---|---|---|
| Anti-JNK1 | 1/1000 | Abcam | 48 kDA | ab110724 |
| Anti-FAK | 1/1000 | Abcam | 125 KDA (119 kDA) | ab40794 |
| Anti-Vinculin | 1/1000 | Abcam | 130 kDA | ab18058 |
| Anti-Caveolin-1 | 1/1000 | Abcam | 22 kDA | ab2910 |
| Anti-NfκB p65 | 1/1000 | Cell-Signaling | 65 kDA | #C22B4 |
| Anti-GAPDH | 1/1000 | Cell-Signaling | 37 kDA | #5174 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bauer, J.; Kopp, S.; Schlagberger, E.M.; Grosse, J.; Sahana, J.; Riwaldt, S.; Wehland, M.; Luetzenberg, R.; Infanger, M.; Grimm, D. Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine. Int. J. Mol. Sci. 2017, 18, 546. https://doi.org/10.3390/ijms18030546
Bauer J, Kopp S, Schlagberger EM, Grosse J, Sahana J, Riwaldt S, Wehland M, Luetzenberg R, Infanger M, Grimm D. Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine. International Journal of Molecular Sciences. 2017; 18(3):546. https://doi.org/10.3390/ijms18030546
Chicago/Turabian StyleBauer, Johann, Sascha Kopp, Elisabeth Maria Schlagberger, Jirka Grosse, Jayashree Sahana, Stefan Riwaldt, Markus Wehland, Ronald Luetzenberg, Manfred Infanger, and Daniela Grimm. 2017. "Proteome Analysis of Human Follicular Thyroid Cancer Cells Exposed to the Random Positioning Machine" International Journal of Molecular Sciences 18, no. 3: 546. https://doi.org/10.3390/ijms18030546