Neuropeptide VGF Promotes Maturation of Hippocampal Dendrites That Is Reduced by Single Nucleotide Polymorphisms
Abstract
:1. Introduction
2. Results
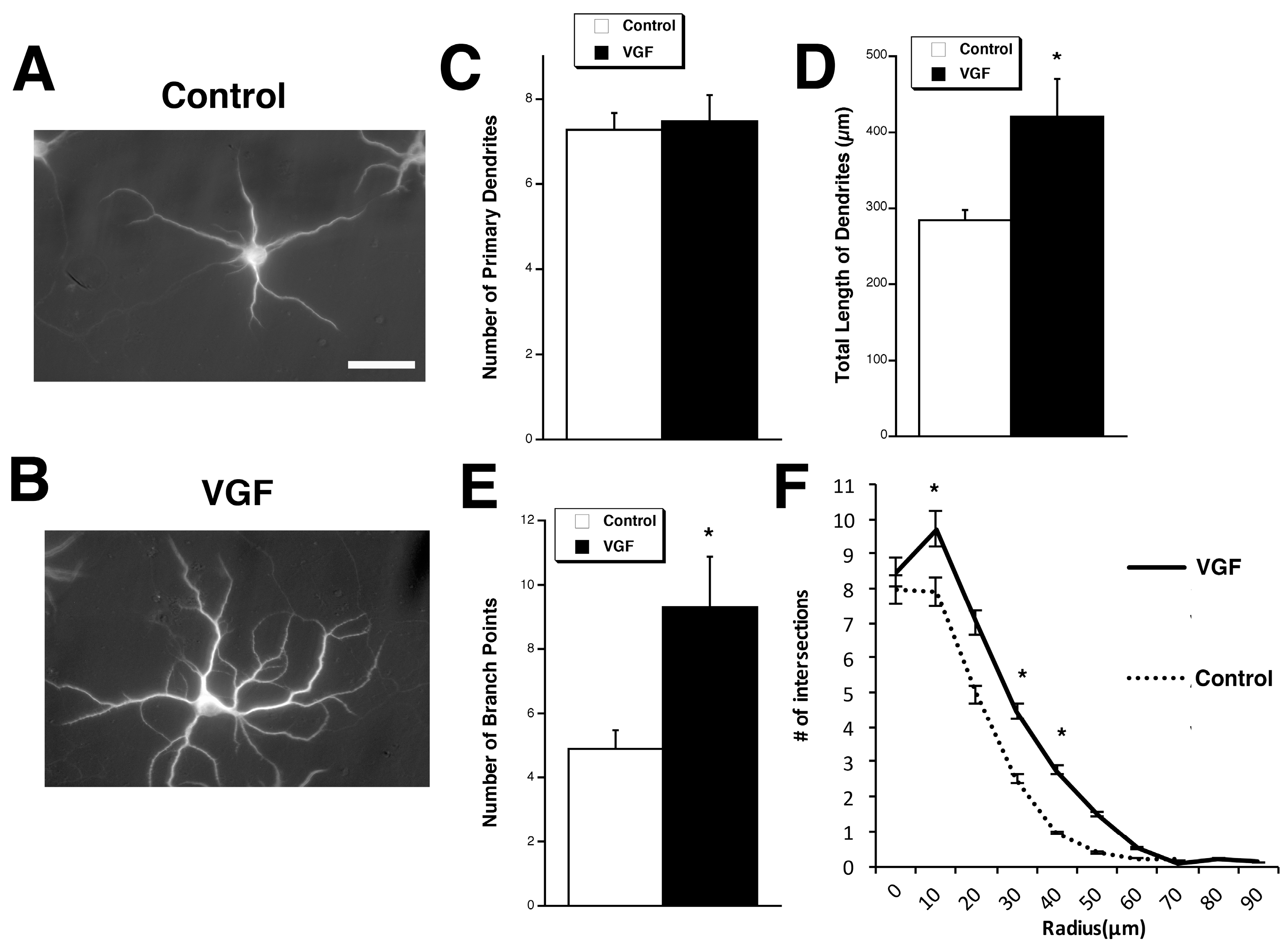
2.1. TLQP-62 Increases Dendritic Branching and Length but Not the Number of Primary Dendrites
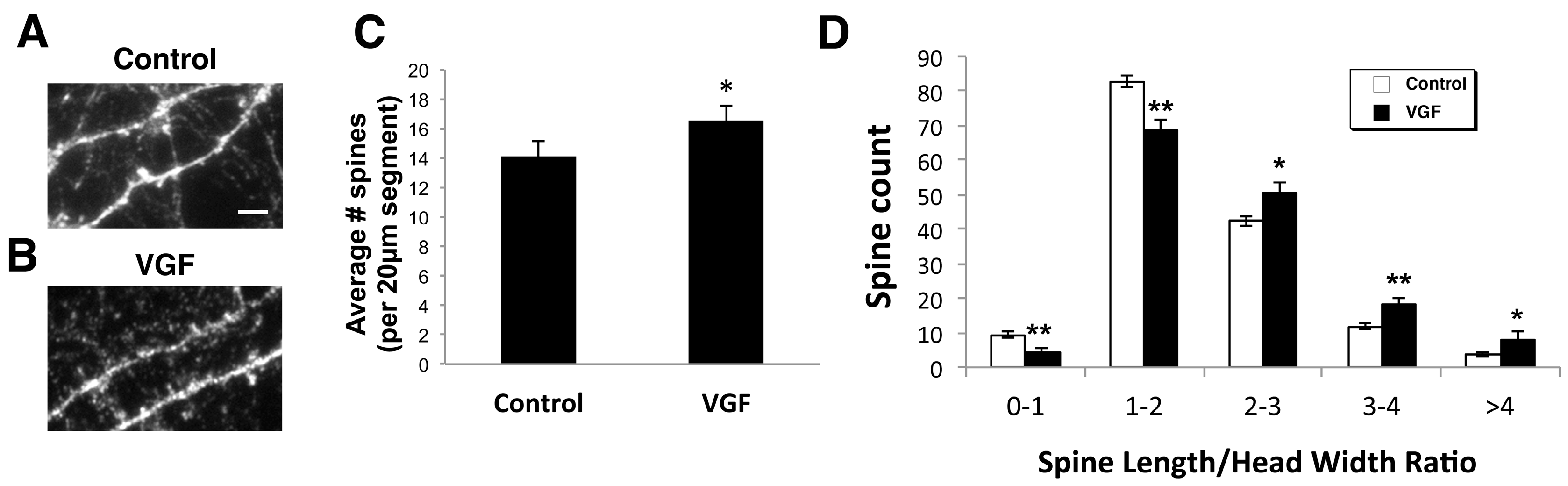
2.2. AAV VGF Increases the Number of Immature Spines in Primary Hippocampal Cultures
2.3. TLQP-62 Peptide Increases Levels of Synaptic Proteins and Number of Functional Synapses In Vitro
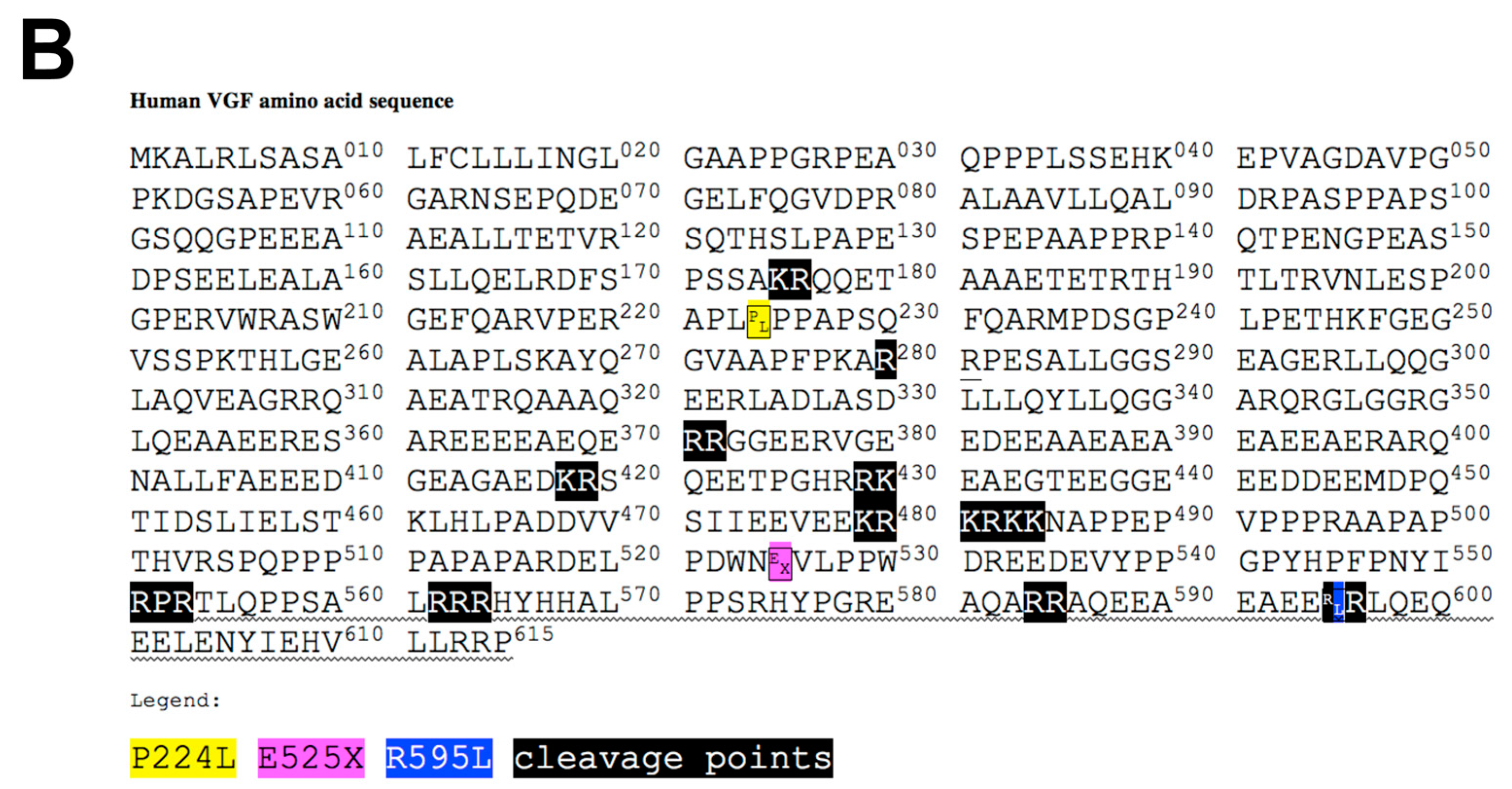
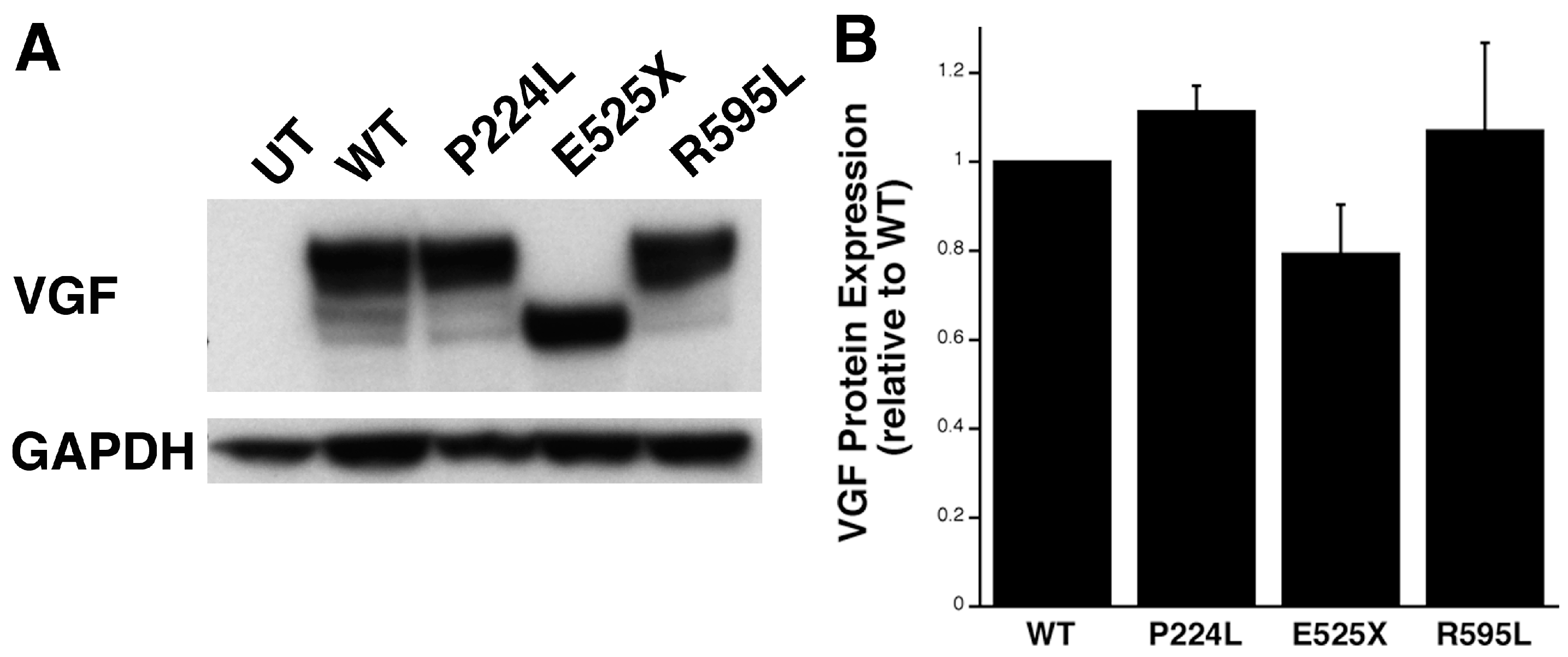
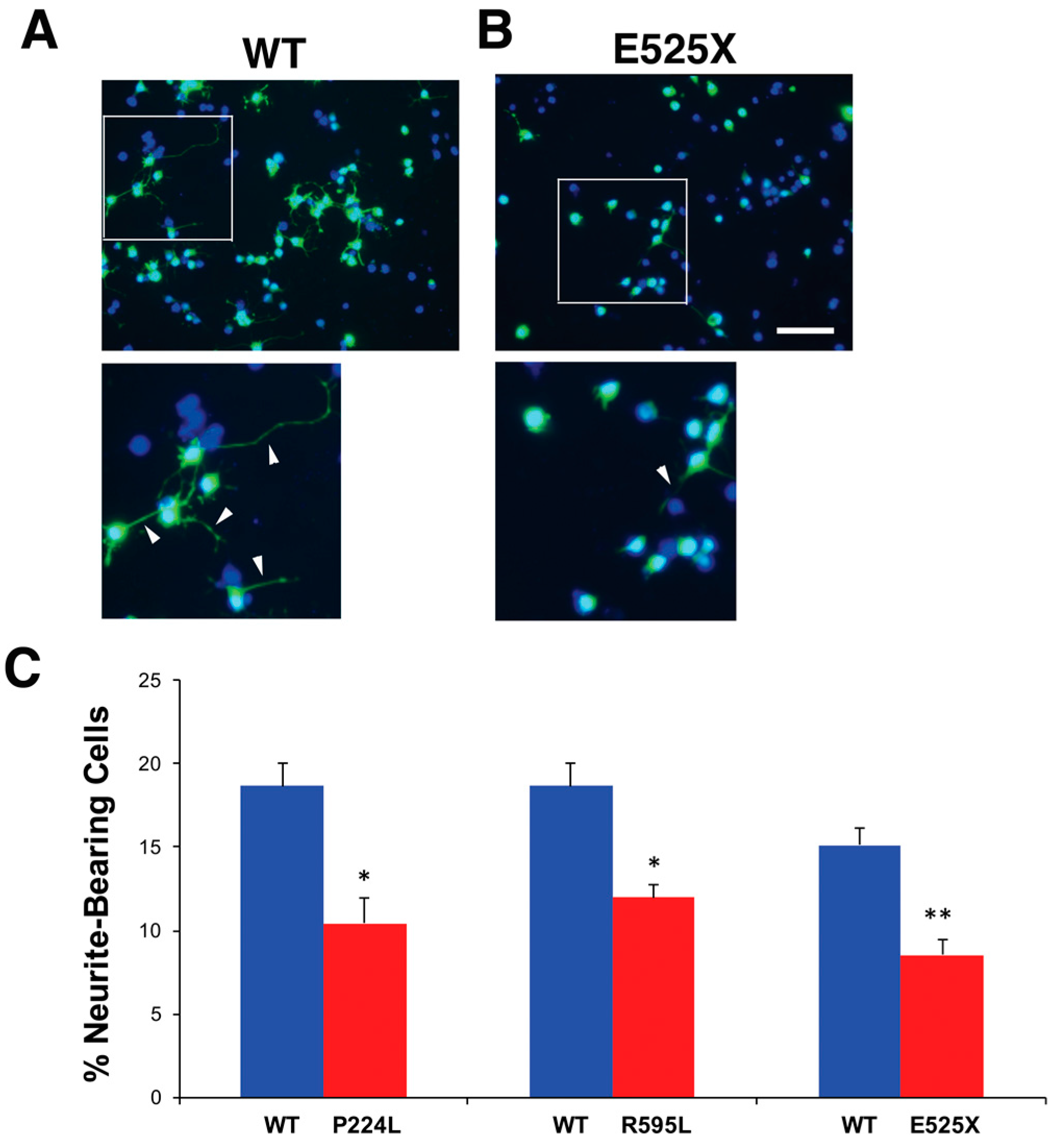
2.4. Identification of Potential Deleterious Non-Synonymous Missense and Nonsense VGF Single Nucleotide Polymorphisms and Their Effects on Protein Expression
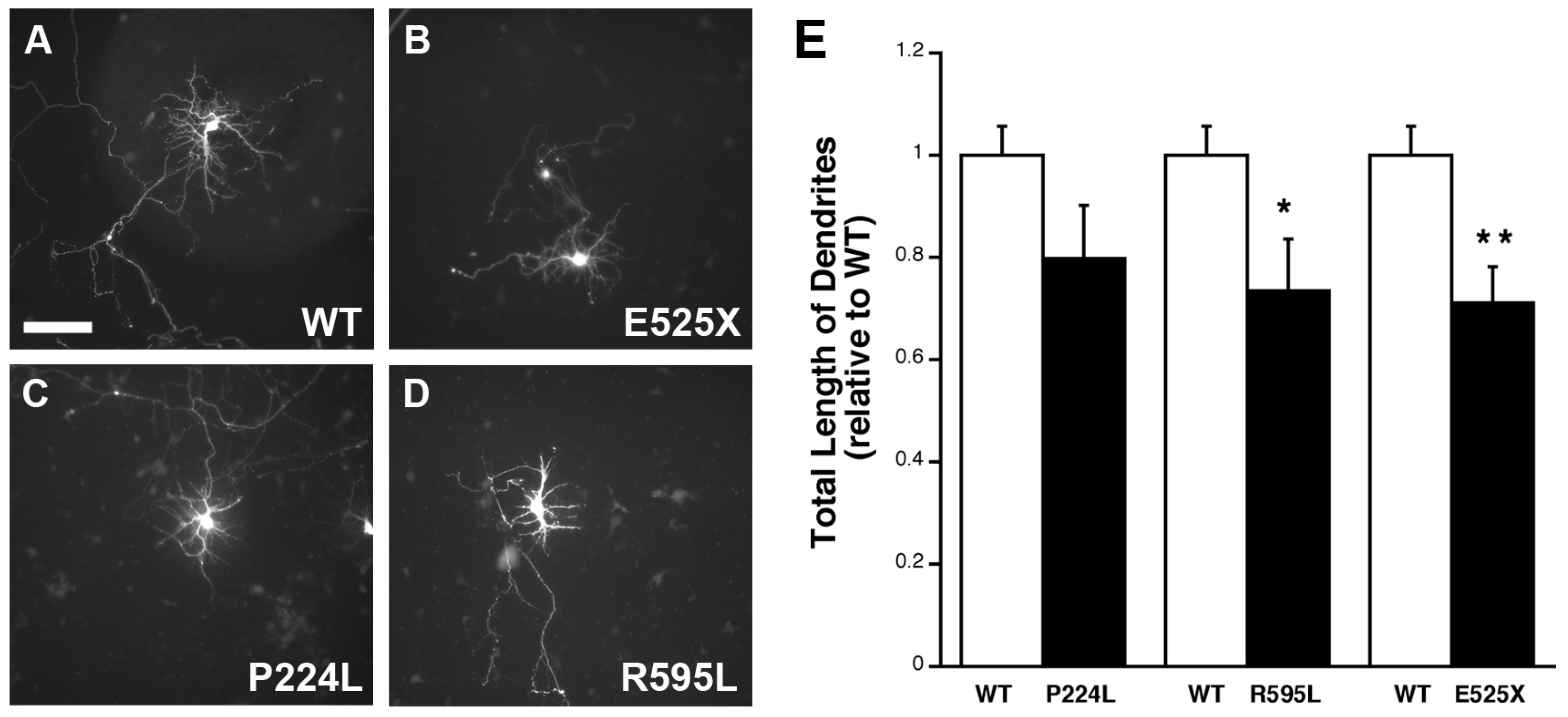
2.5. VGF SNPs Reduce Process Outgrowth in N2a and Primary Hippocampal Cultures
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Hippocampal Cultures
4.3. Dendritic Branching
4.4. Dendritic Spines
4.5. Synaptic Protein Immunostaining
4.6. Synaptic Protein Western Blots
4.7. SNP Structure and Function Prediction
4.8. VGF SNP Plasmid Site-Directed Mutagenesis
4.9. Analysis of SNPs in HEK cells
4.10. SNP Effect on N2A Cell Process Outgrowth
4.11. SNP Effect on Hippocampal Neurite Outgrowth
4.12. Data Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Alder, J.; Thakker-Varia, S.; Bangasser, D.A.; Kuroiwa, M.; Plummer, M.R.; Shors, T.J.; Black, I.B. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J. Neurosci. 2003, 23, 10800–10808. [Google Scholar] [PubMed]
- Trani, E.; Giorgi, A.; Canu, N.; Amadoro, G.; Rinaldi, A.M.; Halban, P.A.; Ferri, G.L.; Possenti, R.; Schinina, M.E.; Levi, A. Isolation and characterization of VGF peptides in rat brain. Role of PC1/3 and PC2 in the maturation of VGF precursor. J. Neurochem. 2002, 81, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; La Corte, G.; Possenti, R.; Locatelli, V.; Rigamonti, A.E.; Torsello, A.; Bresciani, E.; Bulgarelli, I.; Rizzi, R.; Pavone, F.; et al. TLQP-21, a VGF-derived peptide, increases energy expenditure and prevents the early phase of diet-induced obesity. Proc. Natl. Acad. Sci. USA 2006, 103, 14584–14589. [Google Scholar] [CrossRef] [PubMed]
- Hahm, S.; Mizuno, T.M.; Wu, T.J.; Wisor, J.P.; Priest, C.A.; Kozak, C.A.; Boozer, C.N.; Peng, B.; McEvoy, R.C.; Good, P.; et al. Targeted deletion of the VGF gene indicates that the encoded secretory peptide precursor plays a novel role in the regulation of energy balance. Neuron 1999, 23, 537–548. [Google Scholar] [CrossRef]
- Watson, E.; Hahm, S.; Mizuno, T.M.; Windsor, J.; Montgomery, C.; Scherer, P.E.; Mobbs, C.V.; Salton, S.R. VGF ablation blocks the development of hyperinsulinemia and hyperglycemia in several mouse models of obesity. Endocrinology 2005, 146, 5151–5163. [Google Scholar] [CrossRef] [PubMed]
- Foglesong, G.D.; Huang, W.; Liu, X.; Slater, A.M.; Siu, J.; Yildiz, V.; Salton, S.R.; Cao, L. Role of Hypothalamic VGF in Energy Balance and Metabolic Adaption to Environmental Enrichment in Mice. Endocrinology 2016, 157, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Sadahiro, M.; Erickson, C.; Lin, W.J.; Shin, A.C.; Razzoli, M.; Jiang, C.; Fargali, S.; Gurney, A.; Kelley, K.A.; Buettner, C.; et al. Role of VGF-derived carboxy-terminal peptides in energy balance and reproduction: Analysis of "humanized" knockin mice expressing full-length or truncated VGF. Endocrinology 2015, 156, 1724–1738. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.; Ingram, R.; Koch, S.; Theodorou, A.; Low, L.; Baccei, M.; Hathway, G.J.; Costigan, M.; Salton, S.R.; Fitzgerald, M. Origins, actions and dynamic expression patterns of the neuropeptide VGF in rat peripheral and central sensory neurones following peripheral nerve injury. Mol. Pain 2008, 4, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riedl, M.S.; Braun, P.D.; Kitto, K.F.; Roiko, S.A.; Anderson, L.B.; Honda, C.N.; Fairbanks, C.A.; Vulchanova, L. Proteomic analysis uncovers novel actions of the neurosecretory protein VGF in nociceptive processing. J. Neurosci. 2009, 29, 13377–13388. [Google Scholar] [CrossRef] [PubMed]
- Rizzi, R.; Bartolomucci, A.; Moles, A.; D’Amato, F.; Sacerdote, P.; Levi, A.; La Corte, G.; Ciotti, M.T.; Possenti, R.; Pavone, F. The VGF-derived peptide TLQP-21: A new modulatory peptide for inflammatory pain. Neurosci. Lett. 2008, 441, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Pasinetti, G.M.; Salton, S.R. Granins as disease-biomarkers: Translational potential for psychiatric and neurological disorders. Neuroscience 2010, 170, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Cocco, C.; D’Amato, F.; Noli, B.; Ledda, A.; Brancia, C.; Bongioanni, P.; Ferri, G.L. Distribution of VGF peptides in the human cortex and their selective changes in Parkinson’s and Alzheimer’s diseases. J. Anat. 2010, 217, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Shimazawa, M.; Tanaka, H.; Ito, Y.; Morimoto, N.; Tsuruma, K.; Kadokura, M.; Tamura, S.; Inoue, T.; Yamada, M.; Takahashi, H.; et al. An inducer of VGF protects cells against ER stress-induced cell death and prolongs survival in the mutant SOD1 animal models of familial ALS. PLoS ONE 2010, 5, e15307. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Lange, D.J.; Ho, L.; Bonini, S.; Shao, B.; Salton, S.R.; Thomas, S.; Pasinetti, G.M. Vgf is a novel biomarker associated with muscle weakness in amyotrophic lateral sclerosis (ALS), with a potential role in disease pathogenesis. Int. J. Med. Sci. 2008, 5, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Razzoli, M.; Bo, E.; Pascucci, T.; Pavone, F.; D’Amato, F.R.; Cero, C.; Sanghez, V.; Dadomo, H.; Palanza, P.; Parmigiani, S.; et al. Implication of the VGF-derived peptide TLQP-21 in mouse acute and chronic stress responses. Behav. Brain Res. 2012, 229, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.; Rodriguez-Seoane, C.; Rosa, I.; Trossbach, S.V.; Ortega-Alonso, A.; Tomppo, L.; Ekelund, J.; Veijola, J.; Jarvelin, M.R.; Alonso, J.; et al. Neuropeptide precursor VGF is genetically associated with social anhedonia and underrepresented in the brain of major mental illness: Its downregulation by DISC1. Hum. Mol. Genet. 2014, 23, 5859–5865. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Sesta, A.; Calabrese, F.; Nielsen, G.; Riva, M.A.; Gennarelli, M. The Expression of VGF is Reduced in Leukocytes of Depressed Patients and it is Restored by Effective Antidepressant Treatment. Neuropsychopharmacology 2010, 35, 1423–1428. [Google Scholar] [CrossRef] [PubMed]
- Levi, A.; Ferri, G.L.; Watson, E.; Possenti, R.; Salton, S.R. Processing, distribution, and function of VGF, a neuronal and endocrine peptide precursor. Cell. Mol. Neurobiol. 2004, 24, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Snyder, S.E.; Cheng, H.W.; Murray, K.D.; Isackson, P.J.; McNeill, T.H.; Salton, S.R. The messenger RNA encoding VGF, a neuronal peptide precursor, is rapidly regulated in the rat central nervous system by neuronal activity, seizure and lesion. Neuroscience 1998, 82, 7–19. [Google Scholar] [CrossRef]
- Karege, F.; Perret, G.; Bondolfi, G.; Schwald, M.; Bertschy, G.; Aubry, J.M. Decreased serum brain-derived neurotrophic factor levels in major depressed patients. Psychiatry Res. 2002, 109, 143–148. [Google Scholar] [CrossRef]
- Shimizu, E.; Hashimoto, K.; Watanabe, H.; Komatsu, N.; Okamura, N.; Koike, K.; Shinoda, N.; Nakazato, M.; Kumakiri, C.; Okada, S.; et al. Serum brain-derived neurotrophic factor (BDNF) levels in schizophrenia are indistinguishable from controls. Neurosci. Lett. 2003, 351, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Angelucci, F.; Mathe, A.A.; Aloe, L. Neurotrophic factors and CNS disorders: Findings in rodent models of depression and schizophrenia. Prog. Brain Res. 2004, 146, 151–165. [Google Scholar] [PubMed]
- Thakker-Varia, S.; Krol, J.J.; Nettleton, J.; Bilimoria, P.M.; Bangasser, D.A.; Shors, T.J.; Black, I.B.; Alder, J. The neuropeptide VGF produces antidepressant-like behavioral effects and enhances proliferation in the hippocampus. J. Neurosci. 2007, 27, 12156–12167. [Google Scholar] [CrossRef] [PubMed]
- Hunsberger, J.G.; Newton, S.S.; Bennett, A.H.; Duman, C.H.; Russell, D.S.; Salton, S.R.; Duman, R.S. Antidepressant actions of the exercise-regulated gene VGF. Nat. Med. 2007, 13, 1476–1482. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Wang, C.; Xu, B.; Gao, S.; Guo, J.; Zhao, X.; Huang, H.; Zhang, J.; Chen, X.; Wang, Q.; et al. The VGF-derived peptide TLQP62 produces antidepressant-like effects in mice via the BDNF/TrkB/CREB signaling pathway. Pharmacol. Biochem. Behav. 2014, 120, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, C.; Xue, Z.; Li, C.; Zhang, J.; Zhao, X.; Liu, A.; Wang, Q.; Zhou, W. PI3K/AKT/mTOR signaling-mediated neuropeptide VGF in the hippocampus of mice is involved in the rapid onset antidepressant-like effects of GLYX-13. Int. J. Neuropsychopharmacol. 2014, 18. [Google Scholar] [CrossRef] [PubMed]
- Thakker-Varia, S.; Jean, Y.Y.; Parikh, P.; Sizer, C.F.; Jernstedt Ayer, J.; Parikh, A.; Hyde, T.M.; Buyske, S.; Alder, J. The neuropeptide VGF is reduced in human bipolar postmortem brain and contributes to some of the behavioral and molecular effects of lithium. J. Neurosci. 2010, 30, 9368–9380. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Li, M.X.; Xu, C.; Chen, H.B.; An, S.C.; Ma, X.M. Dendritic Spines in Depression: What We Learned from Animal Models. Neural Plast. 2016, 2016, 8056370. [Google Scholar] [CrossRef] [PubMed]
- Neumeister, A.; Wood, S.; Bonne, O.; Nugent, A.C.; Luckenbaugh, D.A.; Young, T.; Bain, E.E.; Charney, D.S.; Drevets, W.C. Reduced hippocampal volume in unmedicated, remitted patients with major depression versus control subjects. Biol. Psychiatry 2005, 57, 935–937. [Google Scholar] [CrossRef] [PubMed]
- Duric, V.; Banasr, M.; Stockmeier, C.A.; Simen, A.A.; Newton, S.S.; Overholser, J.C.; Jurjus, G.J.; Dieter, L.; Duman, R.S. Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects. Int. J. Neuropsychopharmacol. 2013, 16, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Pittenger, C.; Duman, R.S. Stress, depression, and neuroplasticity: A convergence of mechanisms. Neuropsychopharmacology 2008, 33, 88–109. [Google Scholar] [CrossRef] [PubMed]
- Czeh, B.; Simon, M.; Schmelting, B.; Hiemke, C.; Fuchs, E. Astroglial plasticity in the hippocampus is affected by chronic psychosocial stress and concomitant fluoxetine treatment. Neuropsychopharmacology 2006, 31, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- Rocher, C.; Spedding, M.; Munoz, C.; Jay, T.M. Acute stress-induced changes in hippocampal/prefrontal circuits in rats: Effects of antidepressants. Cereb Cortex 2004, 14, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Kellner, Y.; Godecke, N.; Dierkes, T.; Thieme, N.; Zagrebelsky, M.; Korte, M. The BDNF effects on dendritic spines of mature hippocampal neurons depend on neuronal activity. Front. Synaptic Neurosci. 2014, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Lagopoulos, J. Stress and trauma: BDNF control of dendritic-spine formation and regression. Prog. Neurobiol. 2014, 112, 80–99. [Google Scholar] [CrossRef] [PubMed]
- Magarinos, A.M.; Li, C.J.; Gal Toth, J.; Bath, K.G.; Jing, D.; Lee, F.S.; McEwen, B.S. Effect of brain-derived neurotrophic factor haploinsufficiency on stress-induced remodeling of hippocampal neurons. Hippocampus 2011, 21, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Thakker-Varia, S.; Behnke, J.; Doobin, D.; Dalal, V.; Thakkar, K.; Khadim, F.; Wilson, E.; Palmieri, A.; Antila, H.; Rantamaki, T.; et al. VGF (TLQP-62)-induced neurogenesis targets early phase neural progenitor cells in the adult hippocampus and requires glutamate and BDNF signaling. Stem Cell Res. 2014, 12, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, W.; Padhiar, A.; Yue, Y.; Shi, Y.; Zheng, T.; Davis, K.; Zhang, Y.; Huang, M.; Li, Y.; et al. NPAS3 Regulates Transcription and Expression of VGF: Implications for Neurogenesis and Psychiatric Disorders. Front. Mol. Neurosci. 2016, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Mead, B.; Logan, A.; Berry, M.; Leadbeater, W.; Scheven, B.A. Paracrine-mediated neuroprotection and neuritogenesis of axotomised retinal ganglion cells by human dental pulp stem cells: Comparison with human bone marrow and adipose-derived mesenchymal stem cells. PLoS ONE 2014, 9, e109305. [Google Scholar] [CrossRef] [PubMed]
- Severini, C.; Ciotti, M.T.; Biondini, L.; Quaresima, S.; Rinaldi, A.M.; Levi, A.; Frank, C.; Possenti, R. TLQP-21, a neuroendocrine VGF-derived peptide, prevents cerebellar granule cells death induced by serum and potassium deprivation. J. Neurochem. 2008, 104, 534–544. [Google Scholar] [PubMed]
- van den Pol, A.N. Neuropeptide transmission in brain circuits. Neuron 2012, 76, 98–115. [Google Scholar] [CrossRef] [PubMed]
- Altar, C.A.; Laeng, P.; Jurata, L.W.; Brockman, J.A.; Lemire, A.; Bullard, J.; Bukhman, Y.V.; Young, T.A.; Charles, V.; Palfreyman, M.G. Electroconvulsive seizures regulate gene expression of distinct neurotrophic signaling pathways. J. Neurosci. 2004, 24, 2667–2677. [Google Scholar] [CrossRef] [PubMed]
- Hevroni, D.; Rattner, A.; Bundman, M.; Lederfein, D.; Gabarah, A.; Mangelus, M.; Silverman, M.A.; Kedar, H.; Naor, C.; Kornuc, M.; et al. Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J. Mol. Neurosci. 1998, 10, 75–98. [Google Scholar] [CrossRef] [PubMed]
- Newton, S.S.; Collier, E.F.; Hunsberger, J.; Adams, D.; Terwilliger, R.; Selvanayagam, E.; Duman, R.S. Gene profile of electroconvulsive seizures: Induction of neurotrophic and angiogenic factors. J. Neurosci. 2003, 23, 10841–10851. [Google Scholar] [PubMed]
- Bozdagi, O.; Rich, E.; Tronel, S.; Sadahiro, M.; Patterson, K.; Shapiro, M.L.; Alberini, C.M.; Huntley, G.W.; Salton, S.R. The neurotrophin-inducible gene VGF regulates hippocampal function and behavior through a brain-derived neurotrophic factor-dependent mechanism. J. Neurosci. 2008, 28, 9857–9869. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.J.; Jiang, C.; Sadahiro, M.; Bozdagi, O.; Vulchanova, L.; Alberini, C.M.; Salton, S.R. VGF and Its C-Terminal Peptide TLQP-62 Regulate Memory Formation in Hippocampus via a BDNF-TrkB-Dependent Mechanism. J. Neurosci. 2015, 35, 10343–10356. [Google Scholar] [CrossRef] [PubMed]
- Noli, B.; Brancia, C.; Pilleri, R.; D’Amato, F.; Messana, I.; Manconi, B.; Ebling, F.J.; Ferri, G.L.; Cocco, C. Photoperiod Regulates VGF-Derived Peptide Processing in Siberian Hamsters. PLoS ONE 2015, 10, e0141193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amato, F.; Noli, B.; Angioni, L.; Cossu, E.; Incani, M.; Messana, I.; Manconi, B.; Solinas, P.; Isola, R.; Mariotti, S.; et al. VGF Peptide Profiles in Type 2 Diabetic Patients’ Plasma and in Obese Mice. PLoS ONE 2015, 10, e0142333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, H.; Fukutani, Y.; Yamamoto, Y.; Tatara, E.; Takemoto, M.; Shimamura, K.; Yamamoto, N. Thalamus-derived molecules promote survival and dendritic growth of developing cortical neurons. J. Neurosci. 2012, 32, 15388–15402. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Miyazaki, Y.; Kitajo, K.; Yamaguchi, A. VGF, Which Is Induced Transcriptionally in Stroke Brain, Enhances Neurite Extension and Confers Protection Against Ischemia In Vitro. Transl. Stroke Res. 2015, 6, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Pristera, A.; Ayub, M.; Swanwick, R.S.; Karu, K.; Hamada, Y.; Rice, A.S.; Okuse, K. Identification of a receptor for neuropeptide VGF and its role in neuropathic pain. J. Biol. Chem. 2013, 288, 34638–34646. [Google Scholar] [CrossRef] [PubMed]
- Hannedouche, S.; Beck, V.; Leighton-Davies, J.; Beibel, M.; Roma, G.; Oakeley, E.J.; Lannoy, V.; Bernard, J.; Hamon, J.; Barbieri, S.; et al. Identification of the C3a receptor (C3AR1) as the target of the VGF-derived peptide TLQP-21 in rodent cells. J. Biol. Chem. 2013, 288, 27434–27443. [Google Scholar] [CrossRef] [PubMed]
- Peterson, S.L.; Nguyen, H.X.; Mendez, O.A.; Anderson, A.J. Complement protein C1q modulates neurite outgrowth in vitro and spinal cord axon regeneration in vivo. J. Neurosci. 2015, 35, 4332–4349. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Possenti, R.; Mahata, S.K.; Fischer-Colbrie, R.; Loh, Y.P.; Salton, S.R. The extended granin family: Structure, function, and biomedical implications. Endocr. Rev. 2011, 32, 755–797. [Google Scholar] [CrossRef] [PubMed]
- Dijkhuizen, P.A.; Ghosh, A. BDNF regulates primary dendrite formation in cortical neurons via the PI3-kinase and MAP kinase signaling pathways. J. Neurobiol. 2005, 62, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Kawano, Y.; Arimura, N.; Kawabata, S.; Kikuchi, A.; Kaibuchi, K. GSK-3β regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 2005, 120, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Read, D.E.; Gorman, A.M. Involvement of Akt in neurite outgrowth. Cell. Mol. Life Sci. 2009, 66, 2975–2984. [Google Scholar] [CrossRef] [PubMed]
- Tyler, W.J.; Pozzo-Miller, L. Miniature synaptic transmission and BDNF modulate dendritic spine growth and form in rat CA1 neurones. J. Physiol. 2003, 553(Pt. 2), 497–509. [Google Scholar] [CrossRef] [PubMed]
- Castello, N.A.; Nguyen, M.H.; Tran, J.D.; Cheng, D.; Green, K.N.; LaFerla, F.M. 7,8-Dihydroxyflavone, a small molecule TrkB agonist, improves spatial memory and increases thin spine density in a mouse model of Alzheimer disease-like neuronal loss. PLoS ONE 2014, 9, e91453. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Pehrson, A.L.; Waller, J.A.; Dale, E.; Sanchez, C.; Gulinello, M. A critical evaluation of the activity-regulated cytoskeleton-associated protein (Arc/Arg3.1)’s putative role in regulating dendritic plasticity, cognitive processes, and mood in animal models of depression. Front. Neurosci. 2015, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Peebles, C.L.; Yoo, J.; Thwin, M.T.; Palop, J.J.; Noebels, J.L.; Finkbeiner, S. Arc regulates spine morphology and maintains network stability in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 18173–18178. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.L.; Salton, S.R. Expression and polarization of VGF in developing hippocampal neurons. Brain Res. Dev. Brain Res. 1996, 96, 219–228. [Google Scholar] [CrossRef]
- Lombardo, A.; Rabacchi, S.A.; Cremisi, F.; Pizzorusso, T.; Cenni, M.C.; Possenti, R.; Barsacchi, G.; Maffei, L. A developmentally regulated nerve growth factor-induced gene, VGF, is expressed in geniculocortical afferents during synaptogenesis. Neuroscience 1995, 65, 997–1008. [Google Scholar] [CrossRef]
- Colle, R.; Deflesselle, E.; Martin, S.; David, D.J.; Hardy, P.; Taranu, A.; Falissard, B.; Verstuyft, C.; Corruble, E. BDNF/TRKB/P75NTR polymorphisms and their consequences on antidepressant efficacy in depressed patients. Pharmacogenomics 2015, 16, 997–1013. [Google Scholar] [CrossRef] [PubMed]
- Colle, R.; Gressier, F.; Verstuyft, C.; Deflesselle, E.; Lepine, J.P.; Ferreri, F.; Hardy, P.; Guilloux, J.P.; Petit, A.C.; Feve, B.; et al. Brain-derived neurotrophic factor Val66Met polymorphism and 6-month antidepressant remission in depressed Caucasian patients. J. Affect. Disord. 2015, 175, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, R.C.; Lee, A.Y.; Song, Q.; Liaw, A.; Wiener, M.; Paweletz, C.P.; Seeburger, J.L.; Li, J.; Meng, F.; Deyanova, E.G.; et al. High Resolution Discovery Proteomics Reveals Candidate Disease Progression Markers of Alzheimer’s Disease in Human Cerebrospinal Fluid. PLoS ONE 2015, 10, e0135365. [Google Scholar] [CrossRef] [PubMed]
- Jahn, H.; Wittke, S.; Zurbig, P.; Raedler, T.J.; Arlt, S.; Kellmann, M.; Mullen, W.; Eichenlaub, M.; Mischak, H.; Wiedemann, K. Peptide fingerprinting of Alzheimer’s disease in cerebrospinal fluid: Identification and prospective evaluation of new synaptic biomarkers. PLoS ONE 2011, 6, e26540. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.Y.; Hassenstab, J.; Cruchaga, C.; Goate, A.; Fagan, A.M.; Benzinger, T.L.; Maruff, P.; Snyder, P.J.; Masters, C.L.; Allegri, R.; et al. BDNF Val66Met moderates memory impairment, hippocampal function and tau in preclinical autosomal dominant Alzheimer’s disease. Brain 2016, 139(Pt. 10), 2766–2777. [Google Scholar] [CrossRef] [PubMed]
- Wenzler, S.; Knochel, C.; Balaban, C.; Kraft, D.; Kopf, J.; Alves, G.S.; Prvulovic, D.; Carvalho, A.F.; Oertel-Knochel, V. Integrated Biomarkers for Depression in Alzheimer’s Disease: A Critical Review. Curr. Alzheimer Res. 2017, 14, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Musazzi, L.; Treccani, G.; Popoli, M. Functional and structural remodeling of glutamate synapses in prefrontal and frontal cortex induced by behavioral stress. Front. Psychiatry 2015, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.S.; Heninger, G.R.; Nestler, E.J. A molecular and cellular theory of depression. Arch. Gen. Psychiatry 1997, 54, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Sapolsky, R.M. Glucocorticoids and hippocampal atrophy in neuropsychiatric disorders. Arch. Gen. Psychiatry 2000, 57, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Sheline, Y.I.; Wang, P.W.; Gado, M.H.; Csernansky, J.G.; Vannier, M.W. Hippocampal atrophy in recurrent major depression. Proc. Natl. Acad. Sci. USA 1996, 93, 3908–3913. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.S. Neurobiology of stress, depression, and rapid acting antidepressants: Remodeling synaptic connections. Depression Anxiety 2014, 31, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, A.; Gennarelli, M.; Uher, R.; Breen, G.; Farmer, A.; Aitchison, K.J.; Craig, I.W.; Anacker, C.; Zunsztain, P.A.; McGuffin, P.; et al. Candidate genes expression profile associated with antidepressants response in the GENDEP study: Differentiating between baseline ‘predictors’ and longitudinal ‘targets’. Neuropsychopharmacology 2013, 38, 377–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAvoy, K.; Russo, C.; Kim, S.; Rankin, G.; Sahay, A. Fluoxetine induces input-specific hippocampal dendritic spine remodeling along the septotemporal axis in adulthood and middle age. Hippocampus 2015, 25, 1429–1446. [Google Scholar] [CrossRef] [PubMed]
- Duman, C.H.; Duman, R.S. Spine synapse remodeling in the pathophysiology and treatment of depression. Neurosci. Lett. 2015, 601, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Nibuya, M.; Nestler, E.J.; Duman, R.S. Chronic antidepressant administration increases the expression of cAMP response element binding protein (CREB) in rat hippocampus. J. Neurosci. 1996, 16, 2365–2372. [Google Scholar] [PubMed]
- Park, S.W.; Lee, J.G.; Seo, M.K.; Cho, H.Y.; Lee, C.H.; Lee, J.H.; Lee, B.J.; Baek, J.H.; Seol, W.; Kim, Y.H. Effects of mood-stabilizing drugs on dendritic outgrowth and synaptic protein levels in primary hippocampal neurons. Bipolar Disord. 2015, 17, 278–290. [Google Scholar] [CrossRef] [PubMed]
- Naeve, G.S.; Ramakrishnan, M.; Kramer, R.; Hevroni, D.; Citri, Y.; Theill, L.E. Neuritin: A gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Ring, R.H.; Alder, J.; Fennell, M.; Kouranova, E.; Black, I.B.; Thakker-Varia, S. Transcriptional profiling of brain-derived-neurotrophic factor-induced neuronal plasticity: A novel role for nociceptin in hippocampal neurite outgrowth. J. Neurobiol. 2006, 66, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Leemhuis, J.; Henle, F.; Meyer, D.K. VIP induces the elongation of dendrites and axons in cultured hippocampal neurons: Role of microtubules. Peptides 2007, 28, 1700–1705. [Google Scholar] [CrossRef] [PubMed]
- Pinhasov, A.; Nesher, E.; Gross, M.; Turgeman, G.; Kreinin, A.; Yadid, G. The role of the PACAP signaling system in depression. Curr. Pharm. Des. 2011, 17, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Son, H.; Banasr, M.; Choi, M.; Chae, S.Y.; Licznerski, P.; Lee, B.; Voleti, B.; Li, N.; Lepack, A.; Fournier, N.M.; et al. Neuritin produces antidepressant actions and blocks the neuronal and behavioral deficits caused by chronic stress. Proc. Natl. Acad. Sci. USA 2012, 109, 11378–11383. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Y.; Cline, H.T. Stabilization of dendritic arbor structure in vivo by CaMKII. Science 1998, 279, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Groth, R.D.; Lindskog, M.; Thiagarajan, T.C.; Li, L.; Tsien, R.W. β Ca2+/CaM-dependent kinase type II triggers upregulation of GluA1 to coordinate adaptation to synaptic inactivity in hippocampal neurons. Proc. Natl. Acad. Sci. USA 2011, 108, 828–833. [Google Scholar] [CrossRef] [PubMed]
- Thakker-Varia, S.; Alder, J.; Crozier, R.A.; Plummer, M.R.; Black, I.B. Rab3A is required for brain-derived neurotrophic factor-induced synaptic plasticity: Transcriptional analysis at the population and single-cell levels. J. Neurosci. 2001, 21, 6782–6790. [Google Scholar] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behnke, J.; Cheedalla, A.; Bhatt, V.; Bhat, M.; Teng, S.; Palmieri, A.; Windon, C.C.; Thakker-Varia, S.; Alder, J. Neuropeptide VGF Promotes Maturation of Hippocampal Dendrites That Is Reduced by Single Nucleotide Polymorphisms. Int. J. Mol. Sci. 2017, 18, 612. https://doi.org/10.3390/ijms18030612
Behnke J, Cheedalla A, Bhatt V, Bhat M, Teng S, Palmieri A, Windon CC, Thakker-Varia S, Alder J. Neuropeptide VGF Promotes Maturation of Hippocampal Dendrites That Is Reduced by Single Nucleotide Polymorphisms. International Journal of Molecular Sciences. 2017; 18(3):612. https://doi.org/10.3390/ijms18030612
Chicago/Turabian StyleBehnke, Joseph, Aneesha Cheedalla, Vatsal Bhatt, Maysa Bhat, Shavonne Teng, Alicia Palmieri, Charles Christian Windon, Smita Thakker-Varia, and Janet Alder. 2017. "Neuropeptide VGF Promotes Maturation of Hippocampal Dendrites That Is Reduced by Single Nucleotide Polymorphisms" International Journal of Molecular Sciences 18, no. 3: 612. https://doi.org/10.3390/ijms18030612





