In Vitro and In Vivo Studies on the Structural Organization of Chs3 from Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Results
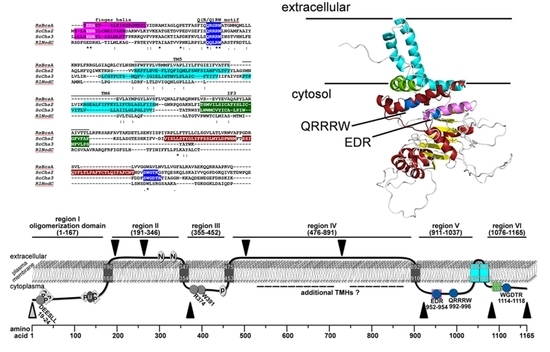
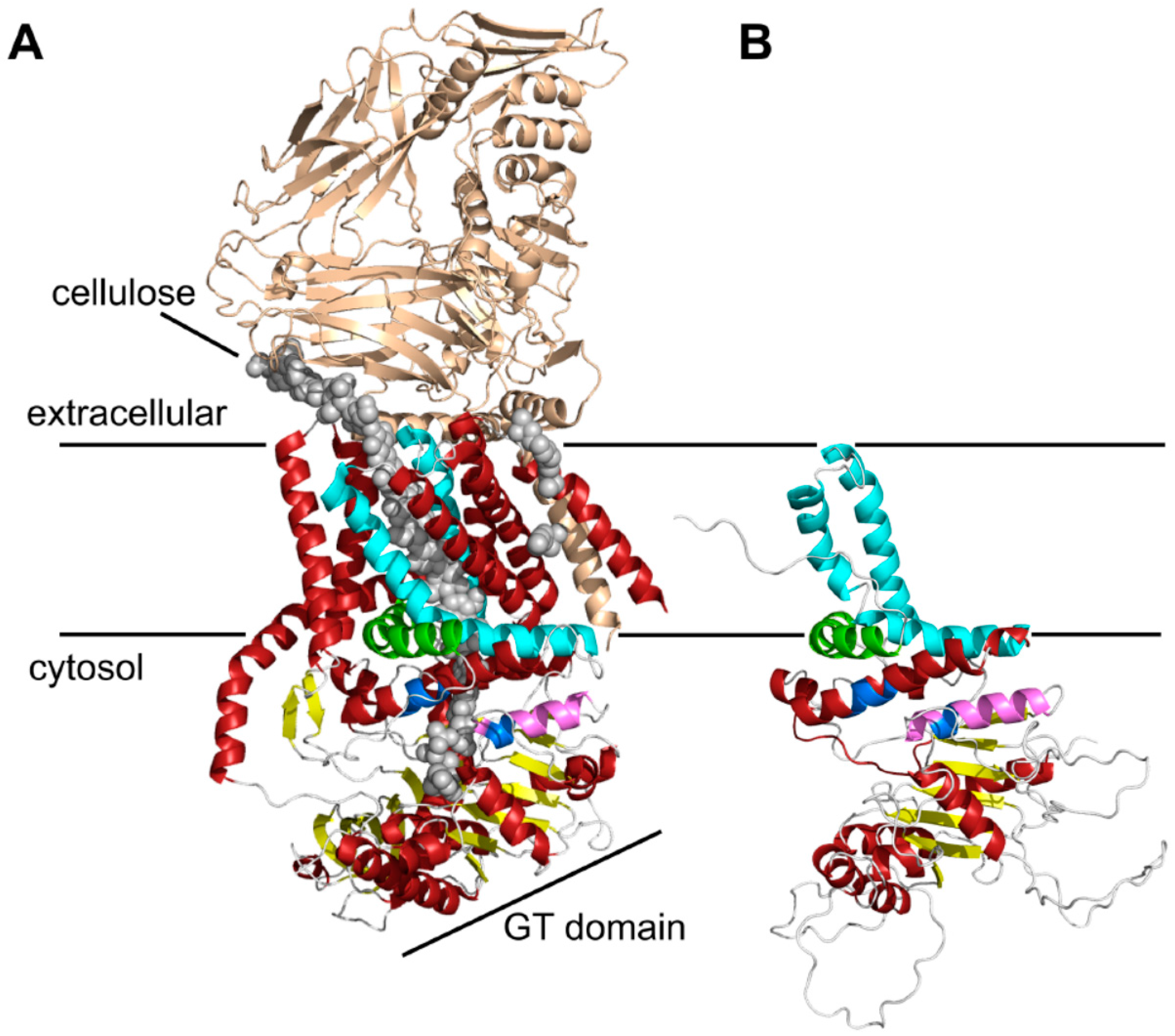
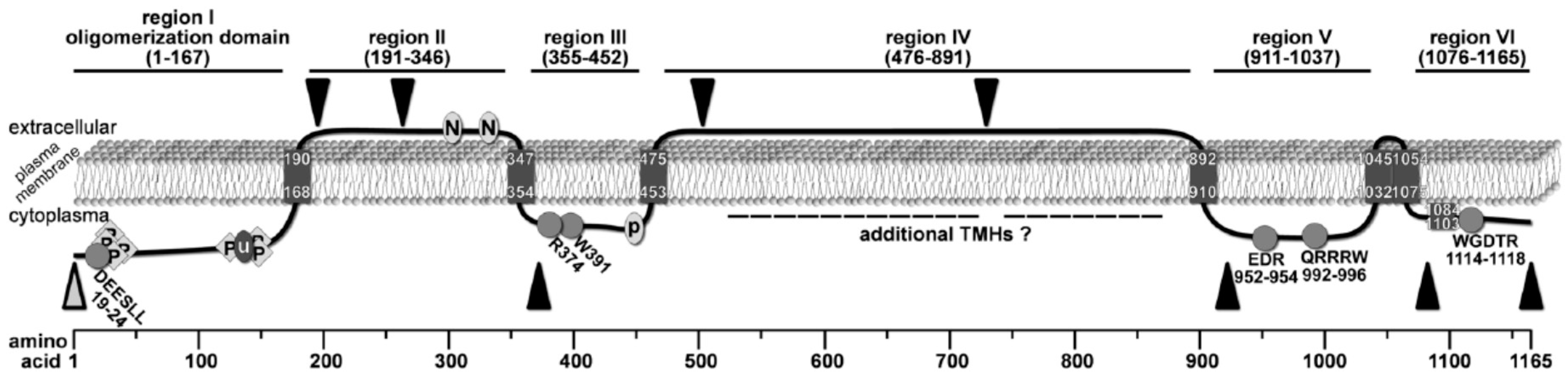
2.1. Bioinformatic Analyses to Predict Membrane Topology and Three-Dimensional (3D) Structure Of Chs3 Reveals Gatekeeper and Finger Helices
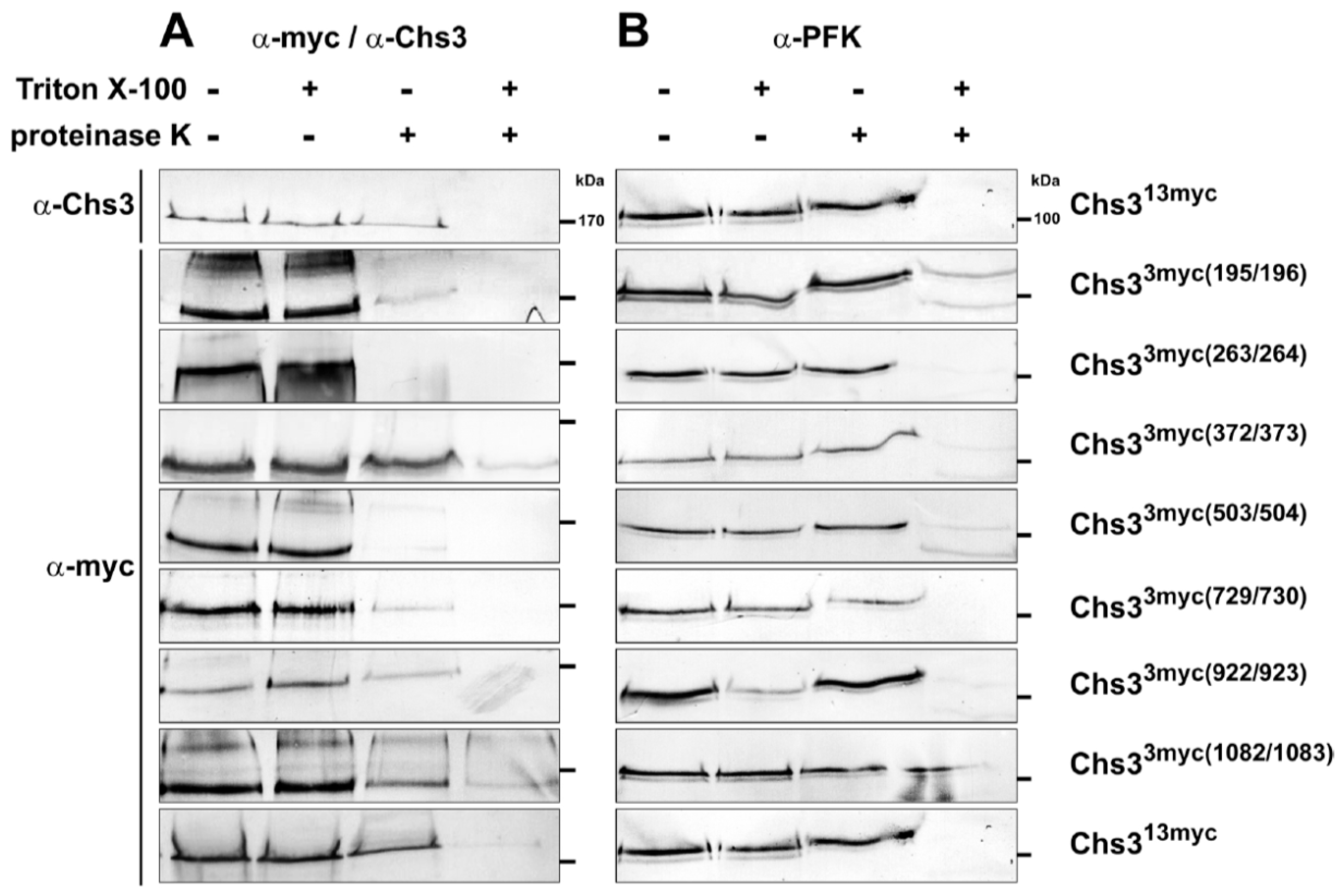
2.2. Protease Protection Assays Suggest a Revised Model on Membrane Topology of Chs3
2.3. Bimolecular Fluorescence Complementation (BiFC) Experiments Demonstrate Di-/Oligomerization of Chs3 In Vivo
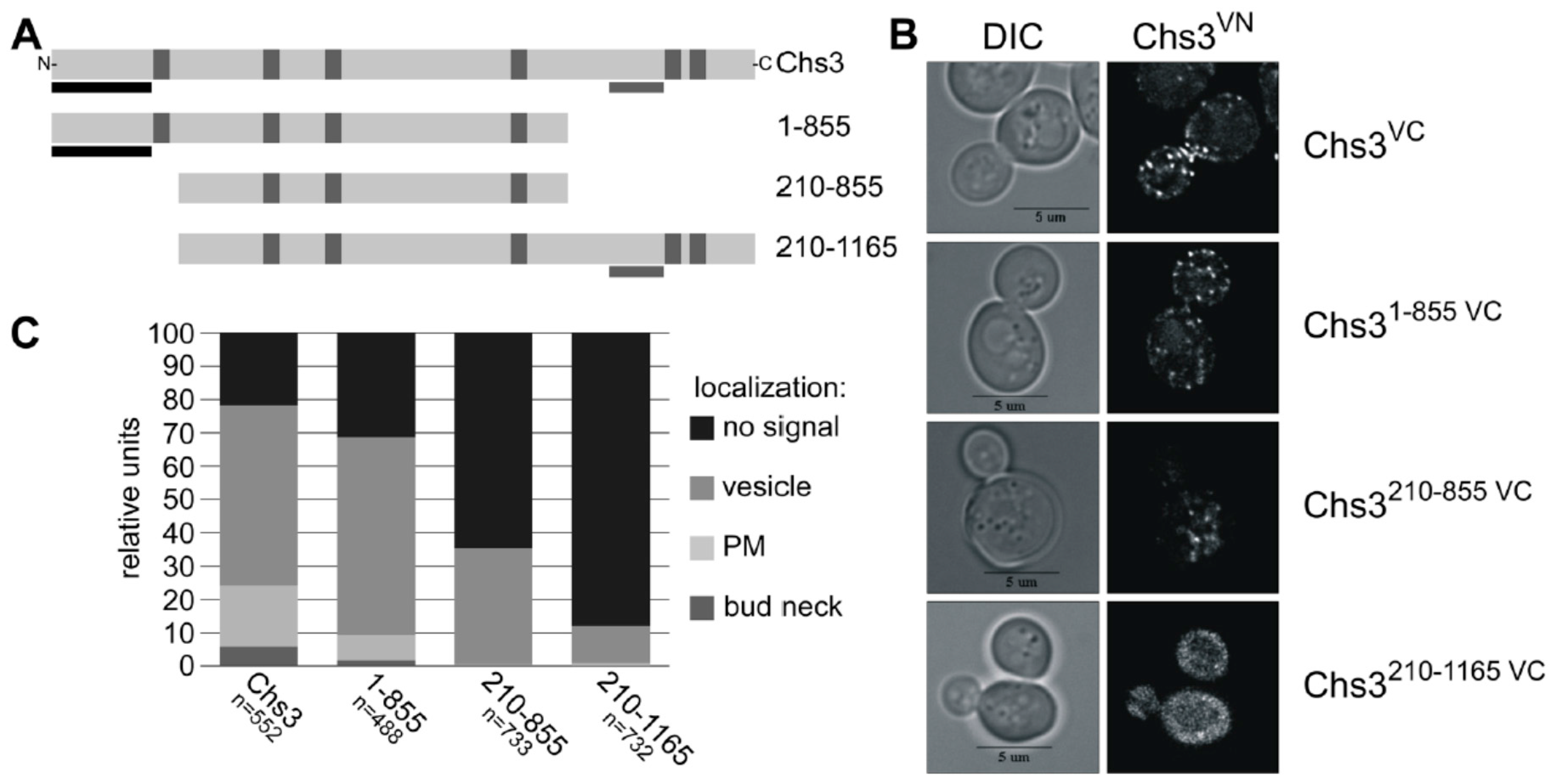
2.4. BiFC Experiments Using Truncated Chs3 Versions Reveal the Site of Self-Interaction
2.5. Tracking Chs3 Di-/Oligomerization in Strains Deficient in Chs3 Trafficking
3. Discussion
4. Experimental Procedures
4.1. Chemicals
4.2. Media
4.3. Construction of Plasmids and Strains
4.4. Complementation Assays in Yeast
4.5. α-Factor Synchronization
4.6. Image Acquisition
4.7. Proteinase K Protection Assays
4.8. Sodium Dodecyl Sulfate Polycarylamide Electrophoresis (SDS-PAGE)/Western Blot
4.9. Bioinformatics
5. Conclusions
Supplementary Materials
Acknowledgements
Author Contributions
Conflicts of Interest
References
- Holan, Z.; Pokorny, V.; Beran, K.; Gemperle, A.; Tuzar, Z.; Baldrian, J. The glucan-chitin complex in Saccharomyces cerevisiae. Arch. Microbiol. 1981, 130, 312–318. [Google Scholar] [CrossRef]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Duran, A. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 2005, 280, 9170–9179. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both β(1–6)- and β(1–3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell 2009, 8, 1626–1636. [Google Scholar] [CrossRef] [PubMed]
- Briza, P.; Ellinger, A.; Winkler, G.; Breitenbach, M. Chemical composition of the yeast ascospore wall. The second outer layer consists of chitosan. J. Biol. Chem. 1988, 263, 11569–11574. [Google Scholar] [PubMed]
- Cabib, E.; Mol, P.C.; Shaw, J.A.; Choi, W.J. Biosynthesis of cell wall and septum during yeast growth. Arch. Med. Res. 1993, 24, 301–303. [Google Scholar] [PubMed]
- Nino-Vega, G.A.; Carrero, L.; San-Blas, G. Isolation of the CHS4 gene of Paracoccidioides brasiliensis and its accommodation in a new class of chitin synthases. Med. Mycol. 2004, 42, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Roncero, C. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 2002, 41, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.S.; Schekman, R.W. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 1996, 135, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, C.P.; Smolka, M.B.; Payne, S.H.; Bafna, V.; Eng, J.; Zhou, H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics 2008, 7, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gerber, S.A.; Rudner, A.D.; Beausoleil, S.A.; Haas, W.; Villen, J.; Elias, J.E.; Gygi, S.P. Large-scale phosphorylation analysis of α-factor-arrested Saccharomyces cerevisiae. J. Proteome Res. 2007, 6, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.K.; Davey, M.; Sun, B.; Roth, A.F.; Davis, N.G.; Conibear, E. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J. Cell Biol. 2006, 174, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Trilla, J.A.; Duran, A.; Roncero, C. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 1999, 145, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Trautwein, M.; Schindler, C.; Gauss, R.; Dengjel, J.; Hartmann, E.; Spang, A. Arf1p, Chs5p and the CHAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006, 25, 943–954. [Google Scholar] [CrossRef] [PubMed]
- Sanchatjate, S.; Schekman, R. Chs5/6 complex: A multiprotein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol. Biol. Cell 2006, 17, 4157–4166. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W.; Hamamoto, S.; Orci, L.; Schekman, R. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 2006, 174, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Ziman, M.; Chuang, J.S.; Tsung, M.; Hamamoto, S.; Schekman, R. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 1565–1576. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.; Castrejon, F.; Duran, A.; Roncero, C. Saccharomyces cerevisiae Bni4p directs the formation of the chitin ring and also participates in the correct assembly of the septum structure. Microbiology 2004, 150, 3229–3241. [Google Scholar] [CrossRef] [PubMed]
- Kozubowski, L.; Panek, H.; Rosenthal, A.; Bloecher, A.; DeMarini, D.J.; Tatchell, K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 2003, 14, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Ziman, M.; Chuang, J.S.; Schekman, R.W. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 1996, 7, 1909–1919. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H. Chitin. In The Sugar Code: Fundamentals of Glycosciences; Gabius, H.-J., Ed.; Wiley-VCH: Weinheim, Germany, 2009; pp. 217–229. [Google Scholar]
- Merzendorfer, H. Insect chitin synthases: A review. J. Comp. Physiol. B 2006, 176, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.L.; Strumillo, J.; Zimmer, J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2013, 493, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Dorfmueller, H.C.; Ferenbach, A.T.; Borodkin, V.S.; van Aalten, D.M. A structural and biochemical model of processive chitin synthesis. J. Biol. Chem. 2014, 289, 23020–23028. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H. Chitin synthesis inhibitors: Old molecules and new developments. Insect Sci. 2013, 20, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Giddings, T.H., Jr.; Brower, D.L.; Staehelin, L.A. Visualization of particle complexes in the plasma membrane of Micrasterias denticulata associated with the formation of cellulose fibrils in primary and secondary cell walls. J. Cell Biol. 1980, 84, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.C.; Brown, R.M., Jr. Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 1980, 84, 315–326. [Google Scholar] [CrossRef] [PubMed]
- DeMarini, D.J.; Adams, A.E.; Fares, H.; de Virgilio, C.; Valle, G.; Chuang, J.S.; Pringle, J.R. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 1997, 139, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Meissner, D.; Odman-Naresh, J.; Vogelpohl, I.; Merzendorfer, H. A novel role of the yeast CaaX protease Ste24 in chitin synthesis. Mol. Biol. Cell 2010, 21, 2425–2433. [Google Scholar] [CrossRef] [PubMed]
- Sacristan, C.; Manzano-Lopez, J.; Reyes, A.; Spang, A.; Muniz, M.; Roncero, C. Oligomerization of the chitin synthase Chs3 is monitored at the Golgi and affects its endocytic recycling. Mol. Microbiol. 2013, 90, 252–266. [Google Scholar] [CrossRef] [PubMed]
- Maue, L.; Meissner, D.; Merzendorfer, H. Purification of an active, oligomeric chitin synthase complex from the midgut of the tobacco hornworm. Insect Biochem. Mol. Biol. 2009, 39, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Cos, T.; Ford, R.A.; Trilla, J.A.; Duran, A.; Cabib, E.; Roncero, C. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 1998, 256, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Yabe, T.; Yamada-Okabe, T.; Nakajima, T.; Sudoh, M.; Arisawa, M.; Yamada-Okabe, H. Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae. Identification of additional amino acid residues involved in its catalytic activity. Eur. J. Biochem. 1998, 258, 941–947. [Google Scholar] [CrossRef]
- Nagahashi, S.; Sudoh, M.; Ono, N.; Sawada, R.; Yamaguchi, E.; Uchida, Y.; Mio, T.; Takagi, M.; Arisawa, M.; Yamada-Okabe, H. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 1995, 270, 13961–13967. [Google Scholar] [CrossRef] [PubMed]
- Sung, M.K.; Huh, W.K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 2007, 24, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Froissard, M.; Belgareh-Touze, N.; Dias, M.; Buisson, N.; Camadro, J.M.; Haguenauer-Tsapis, R.; Lesuisse, E. Trafficking of siderophore transporters in Saccharomyces cerevisiae and intracellular fate of ferrioxamine B conjugates. Traffic 2007, 8, 1601–1616. [Google Scholar] [CrossRef] [PubMed]
- Sacristan, C.; Reyes, A.; Roncero, C. Neck compartmentalization as the molecular basis for the different endocytic behaviour of Chs3 during budding or hyperpolarized growth in yeast cells. Mol. Microbiol. 2012, 83, 1124–1135. [Google Scholar] [CrossRef] [PubMed]
- Fakieh, M.H.; Drake, P.J.; Lacey, J.; Munck, J.M.; Motley, A.M.; Hettema, E.H. Intra-ER sorting of the peroxisomal membrane protein Pex3 relies on its luminal domain. Biol. Open 2013, 2, 829–837. [Google Scholar] [CrossRef] [PubMed]
- Starr, T.L.; Pagant, S.; Wang, C.W.; Schekman, R. Sorting signals that mediate traffic of chitin synthase III between the TGN/endosomes and to the plasma membrane in yeast. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Breidenbach, M.A.; Palaniappan, K.K.; Pitcher, A.A.; Bertozzi, C.R. Mapping yeast N-glycosites with isotopically recoded glycans. Mol. Cell. Proteomics 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Heldermon, C.; DeAngelis, P.L.; Weigel, P.H. Topological organization of the hyaluronan synthase from Streptococcus pyogenes. J. Biol. Chem. 2001, 276, 2037–2046. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Demaeght, P.; Osborne, E.J.; Dermauw, W.; Gohlke, S.; Nauen, R.; Grbic, M.; Tirry, L.; Merzendorfer, H.; Clark, R.M. Population bulk segregant mapping uncovers resistance mutations and the mode of action of a chitin synthesis inhibitor in arthropods. Proc. Natl. Acad. Sci. USA 2012, 109, 4407–4412. [Google Scholar] [CrossRef] [PubMed]
- Demaeght, P.; Osborne, E.J.; Odman-Naresh, J.; Grbic, M.; Nauen, R.; Merzendorfer, H.; Clark, R.M.; van Leeuwen, T. High resolution genetic mapping uncovers chitin synthase-1 as the target-site of the structurally diverse mite growth inhibitors clofentezine, hexythiazox and etoxazole in Tetranychus urticae. Insect Biochem. Mol. Biol. 2014, 51, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Douris, V.; Steinbach, D.; Panteleri, R.; Livadaras, I.; Pickett, J.A.; van Leeuwen, T.; Nauen, R.; Vontas, J. Resistance mutation conserved between insects and mites unravels the benzoylurea insecticide mode of action on chitin biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 14692–14697. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.L.; McNamara, J.T.; Fischer, M.; Rich, J.; Chen, H.M.; Withers, S.G.; Zimmer, J. Observing cellulose biosynthesis and membrane translocation in crystallo. Nature 2016, 531, 329–334. [Google Scholar] [CrossRef] [PubMed]
- McFarlane, H.E.; Doring, A.; Persson, S. The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 2014, 65, 69–94. [Google Scholar] [CrossRef] [PubMed]
- Spang, A. The road not taken: Less traveled roads from the TGN to the plasma membrane. Membranes 2015, 5, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Reyes, A.; Sanz, M.; Duran, A.; Roncero, C. Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane. J. Cell Sci. 2007, 120, 1998–2009. [Google Scholar] [CrossRef] [PubMed]
- Larson, J.R.; Bharucha, J.P.; Ceaser, S.; Salamon, J.; Richardson, C.J.; Rivera, S.M.; Tatchell, K. Protein phosphatase type 1 directs chitin synthesis at the bud neck in Saccharomyces cerevisiae. Mol. Biol. Cell 2008, 19, 3040–3051. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Woods, R.A. High-efficiency transformation of plasmid DNA into yeast. In Molecular Genetics of Yeast; Johnston, J.R., Ed.; Oxford University Press: New York, NY, USA, 1994. [Google Scholar]
- Day, A.; Markwardt, J.; Delaguila, R.; Zhang, J.; Purnapatre, K.; Honigberg, S.M.; Schneider, B.L. Cell size and Cln-Cdc28 complexes mediate entry into meiosis by modulating cell growth. Cell Cycle 2004, 3, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Stoffel, W. TMbase—A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Porollo, A.A.; Adamczak, R.; Meller, J. Polyview: A flexible visualization tool for structural and functional annotations of proteins. Bioinformatics 2004, 20, 2460–2462. [Google Scholar] [CrossRef] [PubMed]
- Kall, L.; Krogh, A.; Sonnhammer, E.L. Advantages of combined transmembrane topology and signal peptide prediction—The phobius web server. Nucleic Acids Res. 2007, 35, W429–W432. [Google Scholar] [CrossRef] [PubMed]
- Cserzo, M.; Eisenhaber, F.; Eisenhaber, B.; Simon, I. TM or not TM: Transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics 2004, 20, 136–137. [Google Scholar] [CrossRef] [PubMed]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Moller, S.; Croning, M.D.; Apweiler, R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 2001, 17, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.; von Heijne, G.; Krogh, A. A hidden markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1998, 6, 175–182. [Google Scholar] [PubMed]
- Reynolds, S.M.; Kall, L.; Riffle, M.E.; Bilmes, J.A.; Noble, W.S. Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput. Biol. 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-based protein structure modeling using the RaptorX web server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gohlke, S.; Muthukrishnan, S.; Merzendorfer, H. In Vitro and In Vivo Studies on the Structural Organization of Chs3 from Saccharomyces cerevisiae. Int. J. Mol. Sci. 2017, 18, 702. https://doi.org/10.3390/ijms18040702
Gohlke S, Muthukrishnan S, Merzendorfer H. In Vitro and In Vivo Studies on the Structural Organization of Chs3 from Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2017; 18(4):702. https://doi.org/10.3390/ijms18040702
Chicago/Turabian StyleGohlke, Simon, Subbaratnam Muthukrishnan, and Hans Merzendorfer. 2017. "In Vitro and In Vivo Studies on the Structural Organization of Chs3 from Saccharomyces cerevisiae" International Journal of Molecular Sciences 18, no. 4: 702. https://doi.org/10.3390/ijms18040702