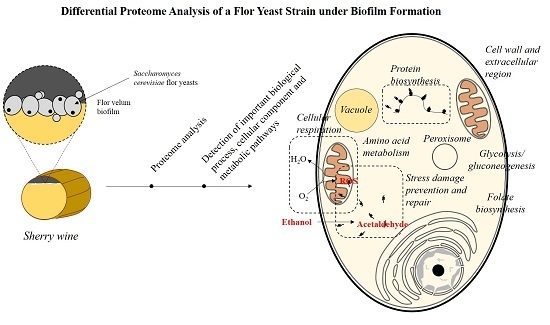
Differential Proteome Analysis of a Flor Yeast Strain under Biofilm Formation
Abstract
:1. Introduction
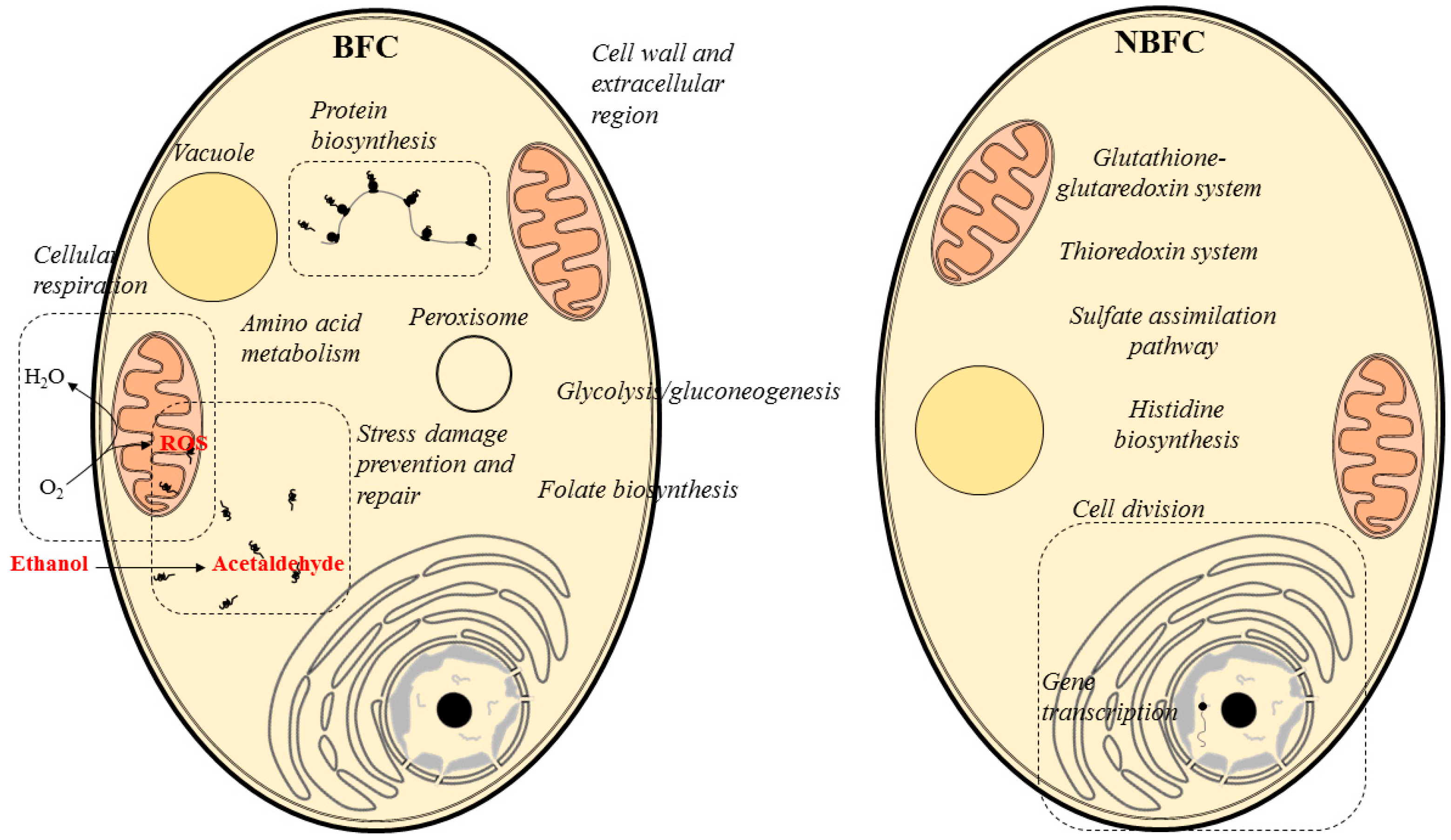
2. Results and Discussion
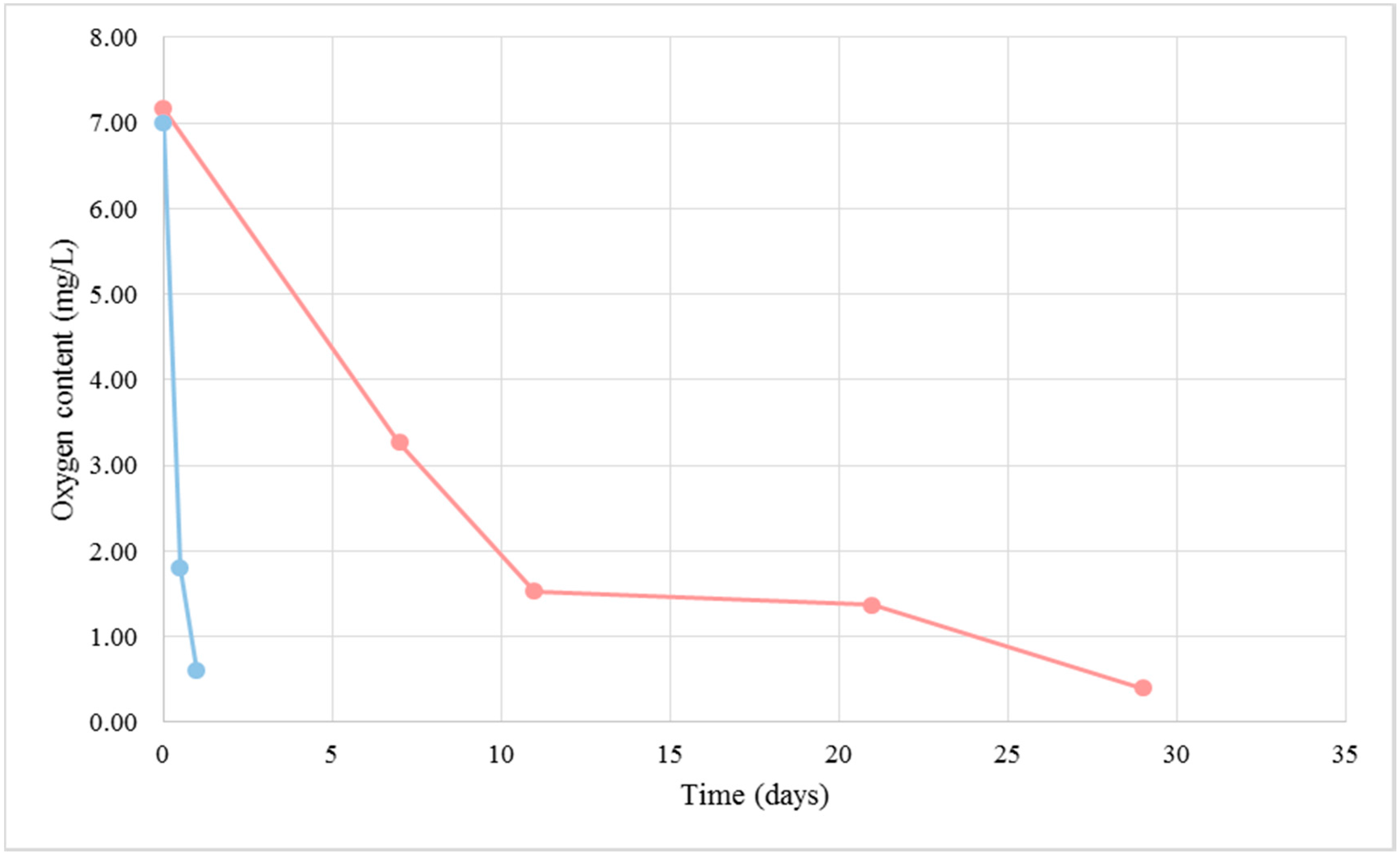
2.1. Cellular Respiration
2.2. Stress Damage Prevention and Repair
2.3. Protein Biosynthesis
2.4. Cell Wall and Extracellular Region
2.5. Peroxisome
2.6. Vacuoles, Proteolysis Regulation and Apoptosis
2.7. Catabolism of Several Sugars
2.8. Amino Acid Metabolism
2.9. Glycolysis/Gluconeogenesis
2.10. Folate Biosynthesis
2.11. Cellular Components, Biological Processes and Pathways of Relevance under NBFC
3. Materials and Methods
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ROS | Reactive oxygen species |
| BFC | Biofilm formation condition |
| NBFC | Non-biofilm formation condition |
| SGD | Saccharomyces Genome Database |
| GO | Gene ontology |
| TCA | Tricarboxylic acid cycle |
| GDC | Glycine decarboxylase complex |
| YPD | Yeast extract, peptone, dextrose medium |
| FDR | False discovery rate |
References
- Peinado, R.A.; Mauricio, J.C. Biologically aged wines. In Wine Chemistry and Biochemistry; Moreno-Arribas, M.V., Polo, M.C., Eds.; Springer: New York, NY, USA, 2009; pp. 81–101. [Google Scholar]
- Alexandre, H. Flor yeasts of Saccharomyces cerevisiae—Their ecology, genetics and metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Castrejon, F.; Codon, A.C.; Cubero, B.; Benıtez, T. Acetaldehyde and ethanol are responsible for mitochondrial DNA restriction fragment length polymorphism in flor yeasts. Syst. Appl. Microbiol. 2002, 25, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Millán, M.C.; Mauricio, J.C. A proteomic and metabolomic approach for understanding the role of the flor yeast mitochondria in the velum formation. Int. J. Food Microbiol. 2014, 172, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Perez-Rodriguez, L.; Benitez, T. Velum formation by flor yeasts isolated from Sherry wines. Am. J. Enol. Vitic. 1997, 48, 55–62. [Google Scholar]
- Ishigami, M.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. FLO11 is essential for flor formation caused by the C–terminal deletion of NRG1 in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2004, 237, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.; Jimenez, J. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233. [Google Scholar] [CrossRef] [PubMed]
- Zara, S.; Farris, A.G.; Budroni, M.; Bakalinsky, A.T. HSP12 is essential for biofilm formation by a Sardinian wine strain of S. cerevisiae. Yeast 2002, 19, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Espinazo-Romeu, M.; Cantoral, J.M.; Matallana, E.; Aranda, A. Btn2p is involved in ethanol tolerance and biofilm formation in flor yeast. FEMS Yeast Res. 2008, 8, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; García-Martínez, T.; Moreno, J.; Mauricio, J.C. Proteins involved in flor yeast carbon metabolism under biofilm formation conditions. Food Microbiol. 2015, 46, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; García-Martínez, T.; Millán, M.C.; Mauricio, J.C.; Moreno, J. Proteins involved in wine aroma compounds metabolism by a Saccharomyces cerevisiae flor-velum yeast strain grown in two conditions. Food Microbiol. 2015, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. Stress responsive proteins of a flor yeast strain during the early stages of biofilm formation. Process Biochem. 2016, 51, 578–588. [Google Scholar] [CrossRef]
- Reddi, A.R.; Culotta, V.C. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell 2013, 152, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, J.C.; Moreno, J.; Ortega, J.M. In vitro specific activities of alcohol and aldehyde dehydrogenases from two flor yeasts during controlled wine aging. J. Agric. Food Chem. 1997, 45, 1967–1971. [Google Scholar] [CrossRef]
- Mauricio, J.C.; Pareja, M.; Ortega, J.M. Changes in the intracellular concentrations of the adenosine phosphates and nicotinamide adenine dinucleotides of Saccharomyces cerevisiae during batch fermentation. World J. Microbiol. Biotechnol. 1995, 11, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Moradas-Ferreira, P.; Costa, V.; Piper, P.; Mager, W. The molecular defences against reactive oxygen species in yeast. Mol. Microbiol. 1996, 19, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Moczko, M.; Schoenfisch, B.; Voos, W.; Pfanner, N.; Rassow, J. The mitochondrial ClpB homolog Hsp78 cooperates with matrix Hsp70 in maintenance of mitochondrial function. J. Mol. Biol. 1995, 254, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Roettgers, K.; Zufall, N.; Guiard, B.; Voos, W. The ClpB homolog Hsp78 is required for the efficient degradation of proteins in the mitochondrial matrix. J. Biol. Chem. 2002, 277, 45829–45837. [Google Scholar] [CrossRef] [PubMed]
- Erjavec, N.; Larsson, L.; Grantham, J.; Nystrom, T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007, 21, 2410–2421. [Google Scholar] [CrossRef] [PubMed]
- Schwelberger, H.G.; Kang, H.A.; Hershey, J.W. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025. [Google Scholar] [PubMed]
- Casey, G.P.; Ingledew, W.M. Ethanol tolerance in yeasts. Crit. Rev. Microbiol. 1986, 13, 219–280. [Google Scholar] [CrossRef] [PubMed]
- Piper, P.W. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 1995, 134, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Nathan, D.F.; Vos, M.H.; Lindquist, S. In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc. Natl. Acad. Sci. USA 1997, 94, 12949–12956. [Google Scholar] [CrossRef] [PubMed]
- Borkovich, K.A.; Farrelly, F.W.; Finkelstein, D.B.; Taulien, J.; Lindquist, S. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell Biol. 1989, 9, 3919–3930. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Gutteridge, J.M.C. Free radicals in Biology and Medicine; Oxford University Press: Oxford, MS, USA, 2015. [Google Scholar]
- Zara, S.; Gross, M.K.; Zara, G.; Budroni, M.; Bakalinsky, A.T. Ethanol–independent biofilm formation by a flor wine yeast strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2010, 76, 4089–4091. [Google Scholar] [CrossRef] [PubMed]
- Higgins, V.J.; Alic, N.; Thorpe, G.W.; Breitenbach, M.; Larsson, V.; Dawes, I.W. Phenotypic analysis of gene deletant strains for sensitivity to oxidative stress. Yeast 2002, 19, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Tanaka, T.; Furusawa, C.; Nagahisa, K.; Hirasawa, T.; Shimizu, H. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 2009, 9, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Ando, A.; Nakamura, T.; Murata, Y.; Takagi, H.; Shima, J. Identification and classification of genes required for tolerance to freeze–thaw stress revealed by genome-wide screening of Saccharomyces cerevisiae deletion strains. FEMS Yeast Res. 2007, 2, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H.; Blanchet, S.; Charpentier, C. Identification of a 49 kDa hydrophobic cell wall mannoprotein present in velum yeast which may be implicated in velum formation. FEMS Microbiol. Lett. 2000, 185, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Pérez Rodríguez, L.; Benítez, T. Factors which affect velum formation by flor yeasts isolated from sherry wine. Syst. Appl. Microbiol. 1997, 20, 154–157. [Google Scholar] [CrossRef]
- Reynolds, T.B.; Fink, G.R. Bakers’ yeast, a model for fungal biofilm formation. Science 2001, 291, 878–881. [Google Scholar] [CrossRef] [PubMed]
- Purevdorj-Gage, B.; Orr, M.E.; Stoodley, P.; Sheehan, K.B.; Hyman, L.E. The role of FLO11 in Saccharomyces cerevisiae biofilm development in a laboratory based flow–cell system. FEMS Yeast Res. 2007, 7, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Zara, S.; Bakalinsky, A.T.; Zara, G.; Pirino, G.; Demontis, M.A.; Budroni, M. FLO11–based model for air–liquid interfacial biofilm formation by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2005, 71, 2934–2939. [Google Scholar] [CrossRef] [PubMed]
- Cullen, P.J.; Sprague, G.F. The regulation of filamentous growth in yeast. Genetics 2012, 190, 23–49. [Google Scholar] [CrossRef] [PubMed]
- Mrsa, V.; Klebl, F.; Tanner, W. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-beta-1,3-glucanase. J. Bacteriol. 1993, 175, 2102–2106. [Google Scholar] [CrossRef] [PubMed]
- Plotnikova, T.A.; Selyakh, I.O.; Kalebina, T.S.; Kulaev, I.S. Bgl2p and Gas1p are the major glucan transferases forming the molecular ensemble of yeast cell wall. Dokl. Biochem. Biophys. 2006, 409, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Klebl, F.; Tanner, W. Molecular cloning of a cell wall exo-beta-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 1989, 171, 6259–6264. [Google Scholar] [CrossRef] [PubMed]
- Tiago, F.C.; Martins, F.S.; Souza, E.L.; Pimenta, P.F.; Araujo, H.R.; Castro, I.M.; Brandao, R.L.; Nicoli, J.R. Adhesion to the yeast cell surface as a mechanism for trapping pathogenic bacteria by Saccharomyces probiotics. J. Med. Microbiol. 2012, 61, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Cappellaro, C.; Mrsa, V.; Tanner, W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 1998, 180, 5030–5037. [Google Scholar] [PubMed]
- Vandenbosch, D.; Canck, E.; Dhondt, I.; Rigole, P.; Nelis, H.; Coenye, T. Genomewide screening for genes involved in biofilm formation and miconazole susceptibility in Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 720–730. [Google Scholar] [CrossRef] [PubMed]
- Destruelle, M.; Holzer, H.; Klionsky, D.J. Identification and characterization of a novel yeast gene: The YGP1 gene product is a highly glycosylated secreted protein that is synthesized in response to nutrient limitation. Mol. Cell. Biol. 1994, 14, 2740–2754. [Google Scholar] [CrossRef] [PubMed]
- Curwin, A.J.; Fairn, G.D.; McMaster, C.R. Phospholipid transfer protein Sec14 is required for trafficking from endosomes and regulates distinct trans-Golgi export pathways. J. Biol. Chem. 2009, 284, 7364–7375. [Google Scholar] [CrossRef] [PubMed]
- Zara, G.; Zara, S.; Pinna, C.; Marceddu, S.; Budroni, M. FLO11 gene length and transcriptional level affect biofilm-forming ability of wild flor strains of Saccharomyces cerevisiae. Microbiology 2009, 159, 3838–3846. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.C.; Raposo, L.R.; Mira, N.P.; Lourenco, A.B.; Sa-Correia, I. Genome–wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol. Appl. Environ. Microbiol. 2009, 75, 5761–5772. [Google Scholar] [CrossRef] [PubMed]
- Zara, G.; Goffrini, P.; Lodi, T.; Zara, S.; Mannazzu, I.; Budroni, M. FLO11 expression and lipid biosynthesis are required for air-liquid biofilm formation in a Saccharomyces cerevisiae flor strain. FEMS Yeast Res. 2012, 12, 864–866. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Kauffman, S.; Reynolds, T.B. Candida albicans uses multiple mechanisms to acquire the essential metabolite inositol during infection. Infect. Immun. 2008, 76, 2793–2801. [Google Scholar] [CrossRef] [PubMed]
- Bethea, E.K.; Carver, B.J.; Montedonico, A.E.; Reynolds, T.B. The inositol regulon controls viability in Candida glabrata. Microbiology 2010, 156, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Mason, D.A.; Shulga, N.; Undavai, S.; Ferrando-May, E.; Rexach, M.F.; Goldfarb, D.S. Increased nuclear envelope permeability and Pep4p-dependent degradation of nucleoporins during hydrogen peroxide-induced cell death. FEMS Yeast Res. 2005, 5, 1237–1251. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Chaves, S.; Alves, S.; Salin, B.; Camougrand, N.; Manon, S.; Joao Sousa, M.; Corte-Real, M. Mitochondrial degradation in acetic acid-induced yeast apoptosis: The role of Pep4 and the ADP/ATP carrier. Mol. Microbiol. 2010, 76, 1398–1410. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, K.U.; Madeo, F. Apoptosis in yeast—A monocellular organism exhibits altruistic behaviour. FEBS Lett. 2000, 473, 6–9. [Google Scholar] [CrossRef]
- Maestre, O.; Santos-Dueñas, I.M.; Peinado, R.; Jiménez-Ot, C.; García-García, I.; Mauricio, J.C. Changes in amino acid composition during wine vinegar production in a fully automatic pilot acetator. Process Biochem. 2008, 43, 803–807. [Google Scholar] [CrossRef]
- Lloyd, D.; Lemar, K.M.; Salgado, L.E.; Gould, T.; Murray, D.B. Respiratory oscillations in yeast: Mitochondrial reactive oxygen species, apoptosis and time, a hypothesis. FEMS Yeast Res. 2003, 3, 333–339. [Google Scholar] [CrossRef]
- Chang, Q.; Petrash, J.M. Disruption of aldo-keto reductase genes leads to elevated markers of oxidative stress and inositol auxotrophy in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2008, 1783, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Garay-Arroyo, A.; Covarrubias, A.A. Three genes whose expression is induced by stress in Saccharomyces cerevisiae. Yeast 1999, 15, 879–892. [Google Scholar] [CrossRef]
- Tkach, J.M.; Yimit, A.; Lee, A.Y.; Riffle, M.; Costanzo, M.; Jaschob, D.; Hendry, J.A.; Ou, J.; Moffat, J.; Boone, C.; et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012, 14, 966–976. [Google Scholar] [CrossRef] [PubMed]
- Magdolen, V.; Oechsner, U.; Trommler, P.; Bandlow, W. Transcriptional control by galactose of a yeast gene encoding a protein homologous to mammalian aldo/keto reductases. Gene 1990, 90, 105–114. [Google Scholar] [CrossRef]
- Kastanos, E.K.; Woldman, Y.Y.; Appling, D.R. Role of mitochondrial and cytoplasmic serine hydroxymethyltransferase isozymes in de novo purine synthesis in Saccharomyces cerevisiae. Biochemistry 1997, 36, 14956–14964. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A.; Dawes, I.W. Genetics of the synthesis of serine from glycine and the utilization of glycine as sole nitrogen source by Saccharomyces cerevisiae. Genetics 1995, 40, 1213–1222. [Google Scholar]
- Ogur, M.; Liu, T.N.; Cheung, I.; Paulavicius, I.; Wales, W.; Mehnert, D.; Blaise, D. “Active” one-carbon generation in Saccharomyces cerevisiae. J. Bacteriol. 1977, 129, 926–933. [Google Scholar] [PubMed]
- Sinclair, D.A.; Hong, S.P.; Dawes, I.W. Specific induction by glycine of the gene for the P-subunit of glycine decarboxylase from Saccharomyces cerevisiae. Mol. Microbiol. 1996, 19, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Ter Schure, E.G.; Flikweert, M.T.; van Dijken, J.P.; Pronk, J.T.; Verrips, C.T. Pyruvate decarboxylase catalyzes decarboxylation of branched–chain 2-oxo acids but is not essential for fusel alcohol production by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1998, 64, 1303–1307. [Google Scholar] [PubMed]
- Botez, M.I. Folate deficiency and neurological disorders in adults. Med. Hypotheses 1976, 2, 135–140. [Google Scholar] [CrossRef]
- Weinstein, S.J.; Hartman, T.J.; Stolzenberg-Solomon, R.; Pietinen, P.; Barrett, M.J.; Taylor, P.R.; Virtamo, J.; Albanes, D. Null association between prostate cancer and serum folate, vitamin B6, vitamin B12, and homocysteine. Cancer Epidemiol. Biomark. Prev. 2003, 12, 1271–1272. [Google Scholar]
- Grant, C.M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001, 3, 533–341. [Google Scholar] [CrossRef]
- Holmgren, A. Thioredoxin and glutaredoxin systems. J. Biol. Chem. 1989, 264, 13963–13966. [Google Scholar] [PubMed]
- SGD Yeastmine. Available online: http://yeastmine.yeastgenome.org/yeastmine/begin.do (accessed on 2 September 2017).
- Uniprot. Available online: http://www.uniprot.org/ (accessed on 2 September 2017).
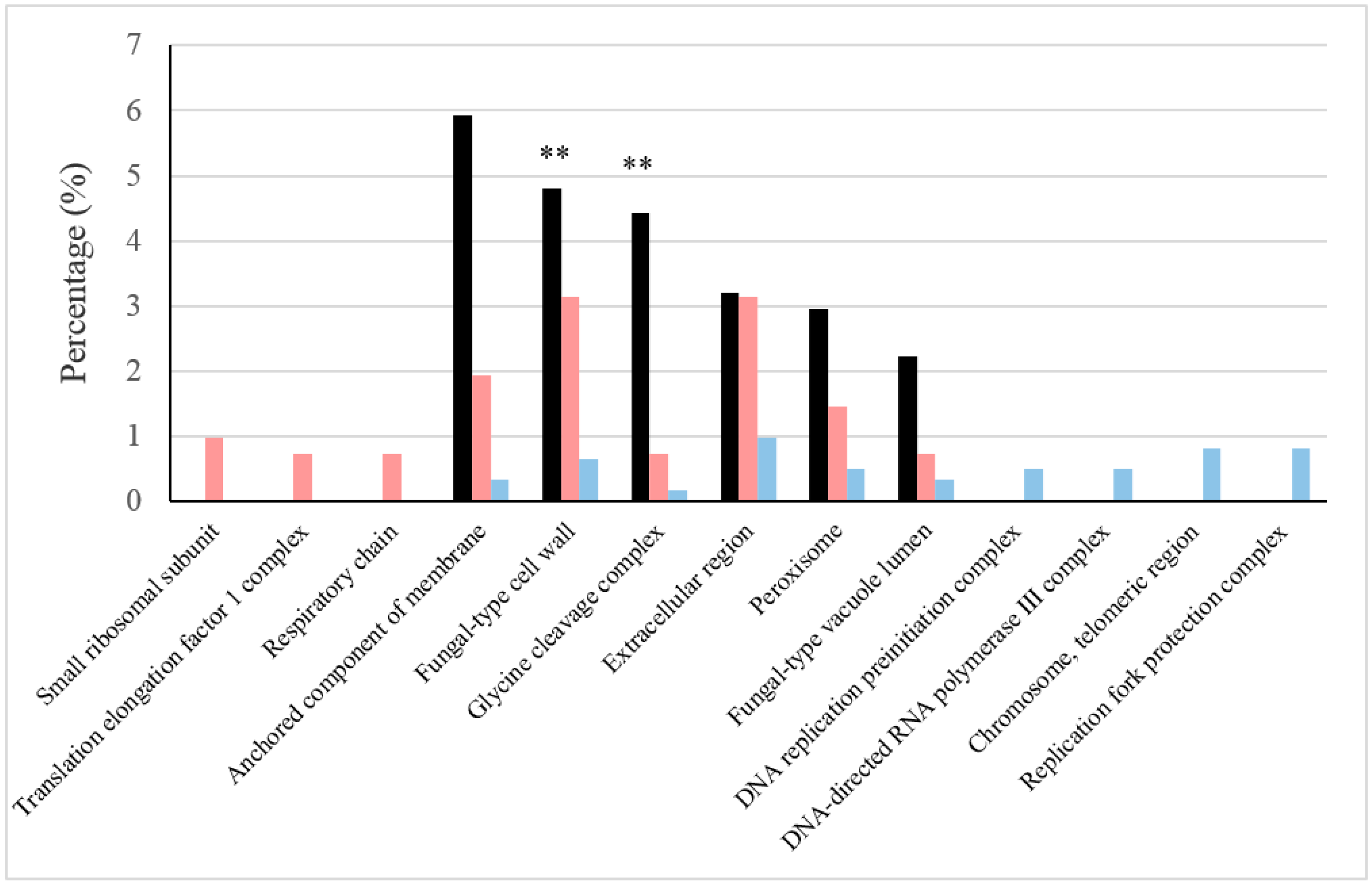

| Cellular Component | BFC Only | BFC and NBFC | NBFC Only |
|---|---|---|---|
| Small ribosomal subunit | Mrp4p, Rps0Bp, Rps15p | Rps0Ap, Rps5p | |
| Translation elongation factor 1 complex | Tef4p | Efb1p, Tef1p | |
| Respiratory chain | Cor1p, Cyb2p | Qcr6p | |
| Anchored component of membrane | Crh1p, Dcw1p, Ecm33p, Plb1p, YJL171C, YNL190W | Ccw14p, Cwp1p, Gas1p, Pst1p | Gas3p, Gas5p |
| Fungal-type cell wall | Bgl2p, Crh1p, Ecm33p, Pho5p, Scw10p, Tos1p, YJL171C, YNL190W | Ccw14p, Cwp1p, Exg1p, Gas1p, Pst1p, Scw4p, Ssa1p, Ssa2p, Tdh1p, Tdh2p, Tdh3p | Gas3p, Gas5p, Hsp150p, Scw11p |
| Glycine cleavage complex | Gcv1p, Gcv3p, Lpd1p | Gcv2p | |
| Extracellular region | Bgl2p, Crh1p, Ecm33p, Pho5p, Scw10p, Suc2p, Tos1p, Ygp1p, YNL190W, YOR389W | Acb1p, Ccw14p, Cwp1p, Exg1p, Gas1p, Pst1p, Pst2p, Scw4p, Ssa1p, Ssa2p | Gas3p, Gas5p, Hsp150p, Scw11p, Uth1p |
| Peroxisome | Cit2p, Hyr1p, Idp3p, Pex1p | Aat2p, Pnc1p | Gpd1p, Pex19p, Pox1p |
| Fungal-type vacuole lumen | Npc2p, Tfs1p | Pep4p | Cps1p, Prb1p |
| DNA replication pre-initiation complex | Mcm7p, Psf1p, Sld5p | ||
| DNA-directed RNA polymerase III complex | Rpc19p, Rpc37p, Rpo26p | ||
| Chromosome, telomeric region | Def1p | Gbp2p, Rap1p, Rfa3p, Rrm3p, Sub2p, | |
| Replication fork protection complex | Ctf4p, Mcm7p, Psf1p, Sld5p, Spt16p |
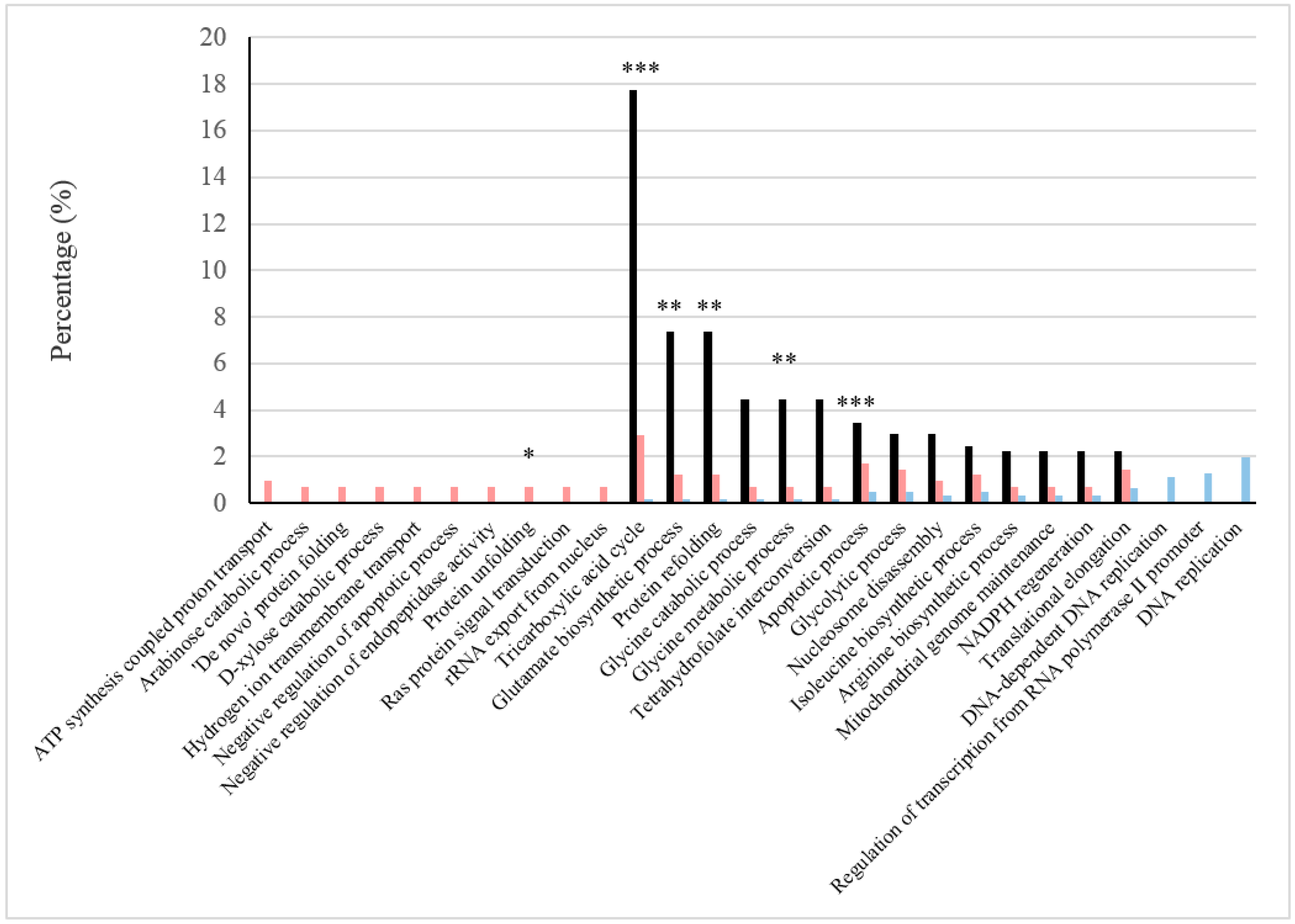
| Biological Processes | BFC Only | BFC and NBFC | NBFC Only |
|---|---|---|---|
| ATP synthesis coupled proton transport | Atp14p, Atp16p, Atp1p, Atp5p | Atp2p | |
| Arabinose catabolic process | Gcy1p, Gre3p | Ypr1p | |
| ‘De novo’ protein folding | Hsp82p, Rot1p, Ydj1p | Hsc82p, Hsp60p | |
| D-xylose catabolic process | Gcy1p, Gre3p | Ypr1p | |
| Hydrogen ion transmembrane transport | Nha1p, Pma1p | Cox4p, Cox6p, Qcr6p | |
| Negative regulation of apoptotic process | Bmh2p | Bmh1p, Stm1p | |
| Negative regulation of endopeptidase activity | Pbi2p, Rfu1p, Tfs1p | ||
| Protein unfolding | Hsp104p, Hsp78p | Ssc1p | |
| Ras protein signal transduction | Bmh2p, Srv2p | Bmh1p | |
| rRNA export from nucleus | Rps0Bp, Rps15p | Rps0Ap, Rps5p | |
| Tricarboxylic acid cycle | Aco1p, Cit1p, Cit2p, Fum1p, Idh1p, Idh2p, Idp3p, Kgd1p, Kgd2p, Mdh1p, Sdh1p | Lsc1p | Aco2p |
| Glutamate biosynthetic process | Cit1p, Cit2p, Idh1p, Idh2p, Put2p | Gdh1p | |
| Protein refolding | Hsp78p, Hsp82p, Ydj1p | Hsc82p, Hsp10p, Hsp60p, Ssa1p, Ssc1p, Sse1p | Mge1p |
| Glycine catabolic process | Gcv1p, Gcv3p, Lpd1p | Gcv2p | |
| Glycine metabolic process | Gcv1p, Shm1p | Shm2p | Gcv2p |
| Tetrahydrofolate interconversion | Shm1p | Ade3p, Shm2p | Met13p |
| Apoptotic process | Bir1p, Kex1p, Mca1p, Pet9p, Por1p | Cpr3p, Tdh2p, Tdh3p | Esp1p, Oye2p, Ymr074cp |
| Glycolytic process | Glk1p, Hxk1p, YLR446W | Cdc19p, Eno1p, Eno2p, Fba1p, Gpm1p, Pdb1p, Pgi1p, Pgk1p, Tdh1p, Tdh2p, Tdh3p, Tpi1p | Hxk2p, Pfk1p |
| Nucleosome disassembly | Asf1pAsf1p, Npl6p, Rsc4p | Rsc8p | Nap1p, Rsc2p |
| Isoleucine biosynthetic process | Hom2p, Ilv3p, Mmf1p | Bat1p, Hom6p, Ilv5p | Bat2p, Ilv2p, Ilv6p |
| Arginine biosynthetic process | Yer069wp | Arg1p, Arg7p, Cpa2p | Arg4p, Arg8p |
| Mitochondrial genome maintenance | Aco1p, Hsp78p, Kgd2p | Ilv5p | Rpo41p, Rrm3p |
| NADPH regeneration | Ald4p, Idp3p, YMR315W | Ald6p | Zwf1p |
| Translational elongation | Tef4p, Tuf1p | Efb1p, Eft1p, Hyp2p, Rpp0p, Rpp2Ap, Rpp2Bp, Rps21Ap, Stm1p, Tef1p | Rpp1Bp, Yef3p, |
| DNA-dependent DNA replication | Abf1p, Ctf4p, Dpb4p, Psf1p, Sld5p, Spt16p, Yil082w-Ap | ||
| Regulation of transcription from RNA polymerase II promoter | Dst1p, Fcp1p, Iki3p, Med4p, Met32p, Ssn2p, Std1p, Ylr278cp | ||
| DNA replication | Pol30p | Abf1p, Ctf4p, Dpb4p, Mcm7p, Pol12p, Psf1p, Rfa3p, Rnr4p, Rrm3p, Sgs1p, Sld5p, Spt16p |
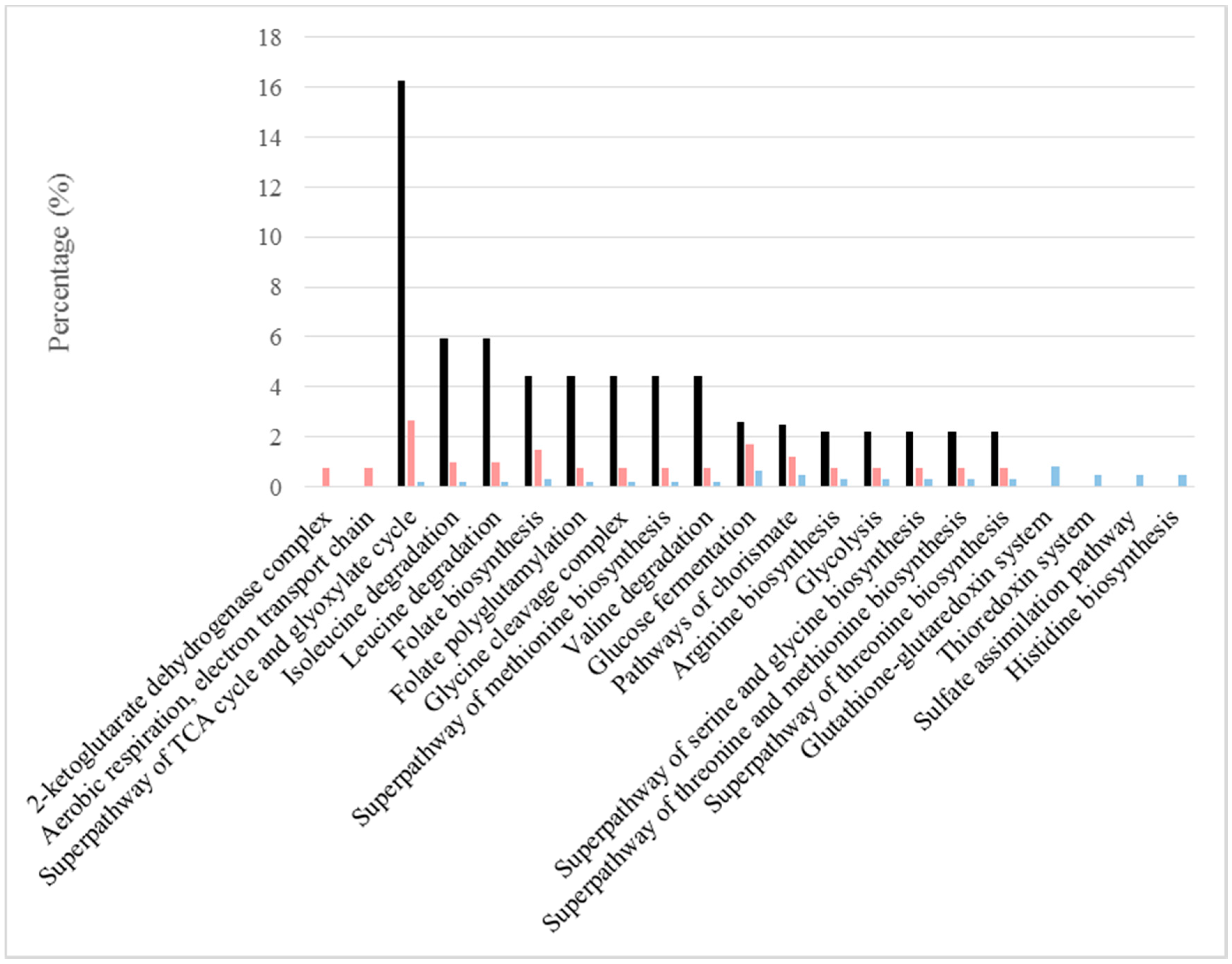
| Pathway | BFC Only | BFC and NBFC | NBFC Only |
|---|---|---|---|
| 2-Ketoglutarate dehydrogenase complex | Kgd1p, Kgd2p, Lpd1p | ||
| Aerobic respiration, electron transport chain | Cor1p, Sdh1p | Cox4p, Cox6p, Qcr6p | |
| Superpathway of TCA cycle and glyoxylate cycle | Aco1p, Cit1p, Cit2p, Fum1p, Idh1p, Idh2p, Kgd1p, Kgd2p, Mdh1p, Sdh1p | Lsc1p | Pyc2p |
| Pathway | BFC Only | BFC and NBFC | NBFC Only |
| Isoleucine degradation | Adh2p, Thi3p | Adh1p, Bat1p, Pdc1p, Pdc5p, Sfa1p | Bat2p |
| Leucine degradation | Adh2p, Thi3p | Adh1p, Bat1p, Sfa1p | Bat2p |
| Folate biosynthesis | Gcv1p, Gcv3p, Lpd1p, Shm1p | Ade3p, Shm2p | Gcv2p, Met13p |
| Folate polyglutamylation | Shm1p | Ade3p, Shm2p | Met13p |
| Glycine cleavage complex | Gcv1p, Gcv3p, Lpd1p | Gcv2p | |
| Superpathway of methionine biosynthesis | Hom2p, Met2p | Hom6p, Met17p, Met6p | |
| Valine degradation | Adh2p | Adh1p, Bat1p, Pdc1p, Pdc5p, Sfa1p | Bat2p |
| Glucose fermentation | Adh2p, Ald4p, Glk1p, Hxk1p | Adh1p, Ald6p, Cdc19p, Eno1p, Eno2p, Fba1p, Gpm1p, Pdc1p, Pdc5p, Pgi1p, Pgk1p, Tdh1p, Tdh2p, Tdh3p, Tpi1p | Hxk2p, Pfk1p, |
| Pathways of chorismate | Aro2p, Shm1p | Ade3p, Aro4p, Shm2p, Trp5p | Aro3p, Aro8p, Trp2p |
| Arginine biosynthesis | Yer069wp | Arg1p, Arg7p, Cpa2p | Arg4p, Arg8p |
| Glycolysis | Cdc19p, Eno1p, Eno2p, Fba1p, Gpm1p, Pgi1p, Pgk1p, Tdh1p, Tdh2p, Tdh3p, Tpi1p | Pfk1p | |
| Superpathway of serine and glycine biosynthesis | Agx1p, Shm1p | Ser1p, Shm2p | Ser33p |
| Superpathway of threonine and methionine biosynthesis | Hom2p, Met2p | Hom6p, Met17p, Met6p | Thr4p |
| Superpathway of threonine biosynthesis | Hom2p | Aat2p, Hom6p | Pyc2p, Thr4p |
| Glutathione-glutaredoxin system | Grx1p, Grx2p | Glr1p, Grx3p, Grx4p, Grx5p | |
| Thioredoxin system | Trx1p, Trx2p | Trr1p | |
| Sulfate assimilation pathway | Ecm17p | Met10p, Met14p | |
| Histidine biosynthesis | His3p, His4p, His7p | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. Differential Proteome Analysis of a Flor Yeast Strain under Biofilm Formation. Int. J. Mol. Sci. 2017, 18, 720. https://doi.org/10.3390/ijms18040720
Moreno-García J, Mauricio JC, Moreno J, García-Martínez T. Differential Proteome Analysis of a Flor Yeast Strain under Biofilm Formation. International Journal of Molecular Sciences. 2017; 18(4):720. https://doi.org/10.3390/ijms18040720
Chicago/Turabian StyleMoreno-García, Jaime, Juan Carlos Mauricio, Juan Moreno, and Teresa García-Martínez. 2017. "Differential Proteome Analysis of a Flor Yeast Strain under Biofilm Formation" International Journal of Molecular Sciences 18, no. 4: 720. https://doi.org/10.3390/ijms18040720