Identification of Novel A2/C2 Inter-Genotype Recombinants of Hepatitis B Virus from a Korean Chronic Patient Co-Infected with Both Genotype A2 and C2
Abstract
:1. Introduction
2. Results
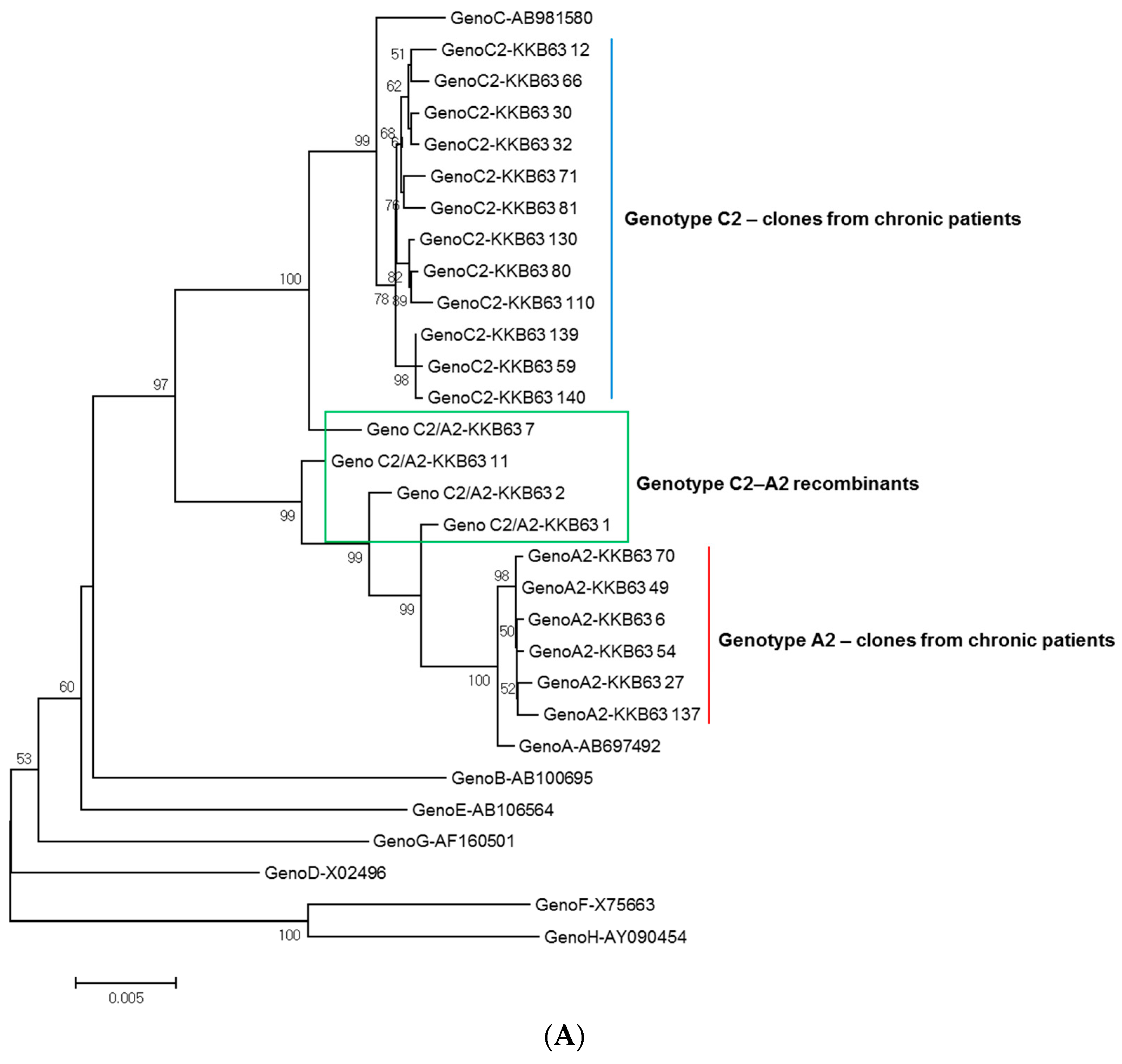
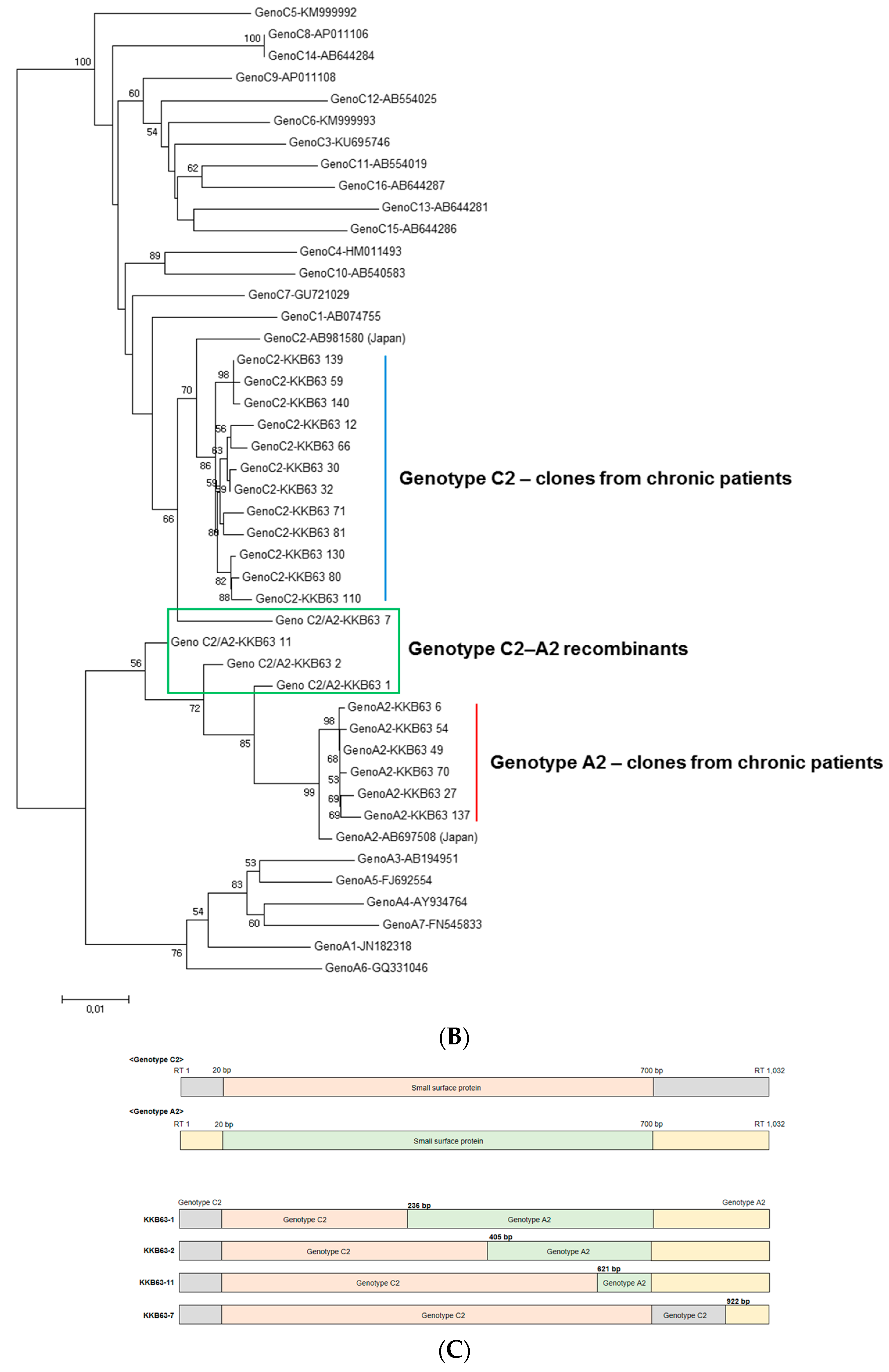
2.1. Identification of Inter-Genotypic Recombinants of Genotype A2/C2 from a Korean Patient, H62, by Phylogenetic Analysis of 1032-bp Polymerase RT Sequences
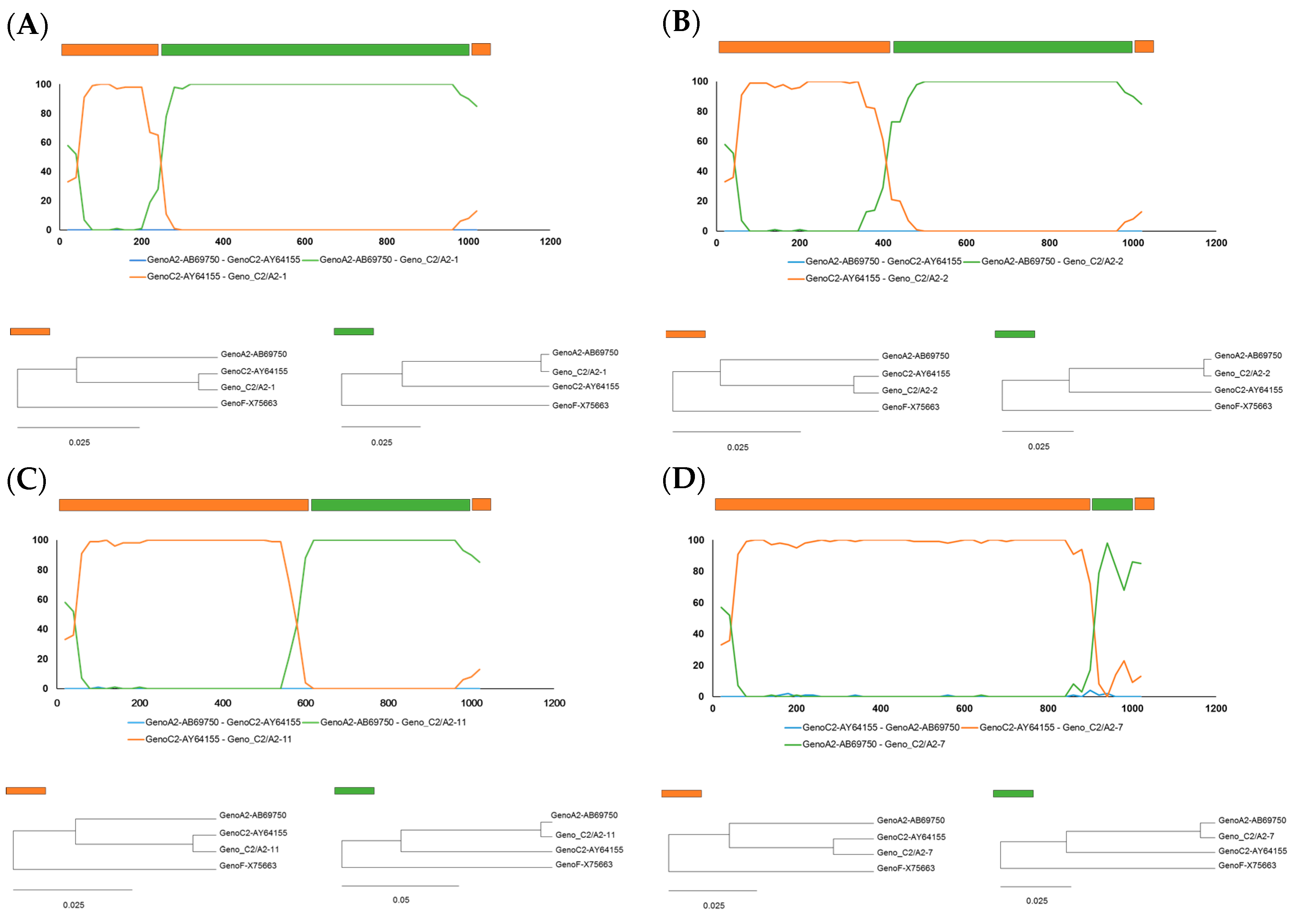
2.2. Identification of Recombinant Breakpoints in Four Recombinant Clones by BootScan Analysis
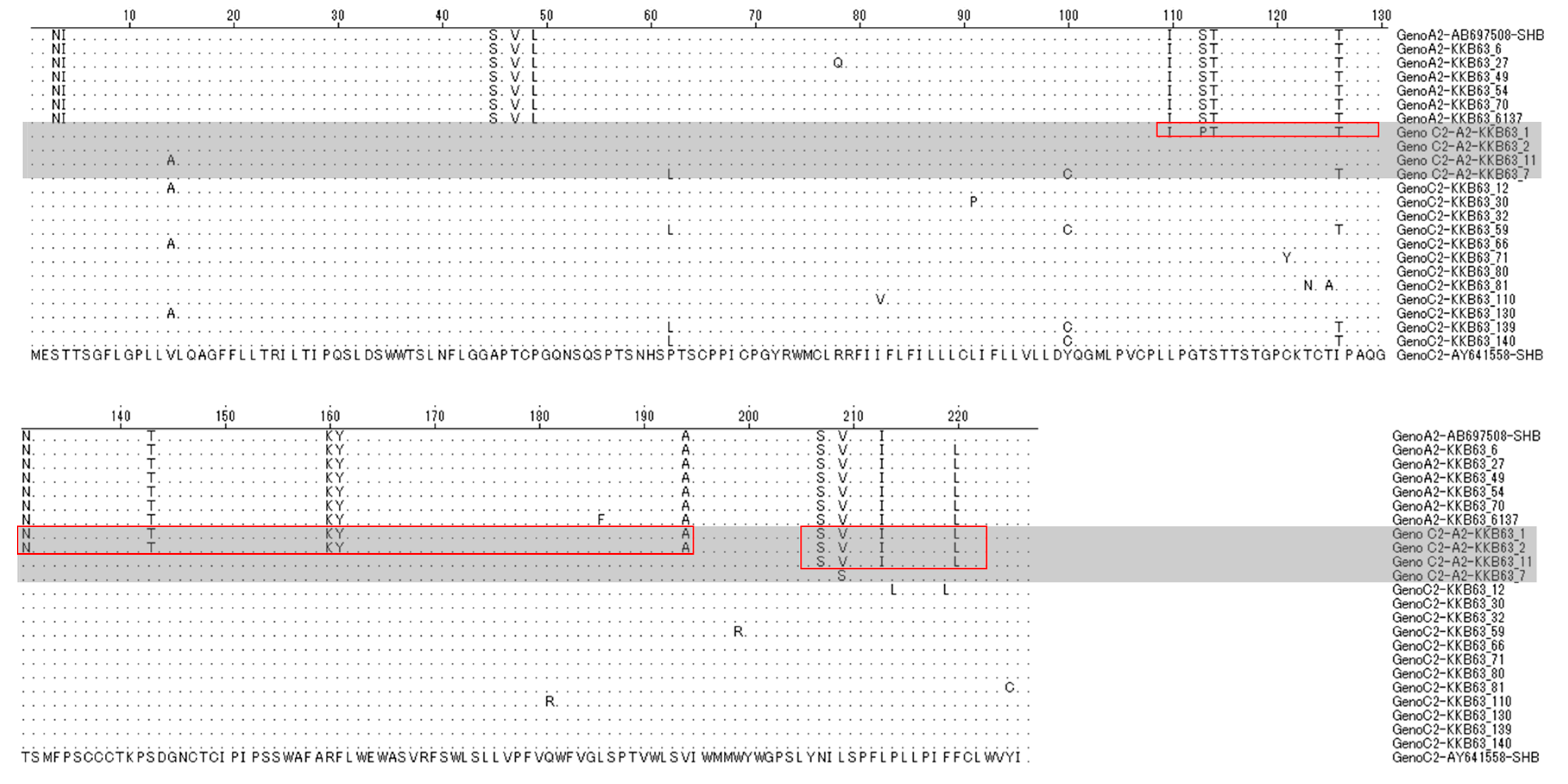
2.3. The Simultaneous Modification of HBsAg by Recombination in the Polymerase RT Region
3. Discussion
4. Materials and Methods
4.1. Patient
4.2. DNA Extraction and Amplification of the Polymerase RT Region
4.3. TA Cloning and Sequencing Analysis of the Amplicon
4.4. HBV Genotyping
4.5. BootScan Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kao, J.H.; Chen, D.S. Global control of hepatitis B virus infection. Lancet Infect. Dis. 2002, 2, 395–403. [Google Scholar] [CrossRef]
- El-Serag, H.B.; Rudolph, K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef] [PubMed]
- Gan, S.I.; Devlin, S.M.; Scott-Douglas, N.W.; Burak, K.W. Lamivudine for the treatment of membranous glomerulopathy secondary to chronic hepatitis B infection. Can. J. Gastroenterol. 2005, 19, 625–629. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 2012, 142, 1264–1273e1. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M. Hepatitis B virus infection. N. Engl. J. Med. 1997, 337, 1733–1745. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, H.; Cho, Y.; Oh, K.; Ki, M. Changes in seroprevalence of hepatitis B surface antigen and epidemiologic characteristics in the Republic of Korea, 1998–2013. Epidemiol. Health 2015, 37, e2015055. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, J.; Rodes, B.; Zoulim, F.; Bartholomeusz, A.; Soriano, V. Mutations affecting the replication capacity of the hepatitis B virus. J. Viral Hepat. 2006, 13, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Torresi, J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J. Clin. Virol. 2002, 25, 97–106. [Google Scholar] [CrossRef]
- Zhang, M.; Ge, G.; Yang, Y.; Cai, X.; Fu, Q.; Cai, J.; Huang, Z. Decreased antigenicity profiles of immune-escaped and drug-resistant hepatitis B surface antigen (HBsAg) double mutants. Virol. J. 2013, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Norder, H.; Hammas, B.; Lofdahl, S.; Courouce, A.M.; Magnius, L.O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992, 73, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Stuyver, L.; De Gendt, S.; Van Geyt, C.; Zoulim, F.; Fried, M.; Schinazi, R.F.; Rossau, R. A new genotype of hepatitis B virus: Complete genome and phylogenetic relatedness. J. Gen. Virol. 2000, 81, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, J.H.; Nam, J.J.; Kim, H.R.; Shin, H.R. Epidemiological findings of hepatitis B infection based on 1998 National Health and Nutrition Survey in Korea. J. Korean Med. Sci. 2002, 17, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Kramvis, A.; Kew, M.C. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J. Viral Hepat. 2005, 12, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Guettouche, T.; Hnatyszyn, H.J. Chronic hepatitis B and viral genotype: The clinical significance of determining HBV genotypes. Antivir. Ther. 2005, 10, 593–604. [Google Scholar] [PubMed]
- Orito, E.; Ichida, T.; Sakugawa, H.; Sata, M.; Horiike, N.; Hino, K.; Okita, K.; Okanoue, T.; Iino, S.; Tanaka, E.; et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 2001, 34, 590–594. [Google Scholar] [CrossRef] [PubMed]
- Norder, H.; Courouce, A.M.; Coursaget, P.; Echevarria, J.M.; Lee, S.D.; Mushahwar, I.K.; Robertson, B.H.; Locarnini, S.; Magnius, L.O. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology 2004, 47, 289–309. [Google Scholar] [CrossRef] [PubMed]
- Kao, J.H.; Chen, P.J.; Lai, M.Y.; Chen, D.S. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J. Clin. Microbiol. 2002, 40, 1207–1209. [Google Scholar] [CrossRef] [PubMed]
- Lavanchy, D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J. Clin. Virol. 2005, 34 (Suppl. S1), S1–S3. [Google Scholar] [CrossRef]
- Kim, H.; Jee, Y.M.; Song, B.C.; Shin, J.W.; Yang, S.H.; Mun, H.S.; Kim, H.J.; Oh, E.J.; Yoon, J.H.; Kim, Y.J.; et al. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology 2007, 50, 52–57. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jee, Y.M.; Song, B.C.; Hyan, J.W.; Mun, H.S.; Kim, H.J.; Oh, E.J.; Yoon, J.H.; Kim, Y.J.; Lee, H.S.; et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J. Med. Virol. 2007, 79, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Park, J.W.; Kim, H.J.; Lee, D.H.; Cha, Y.J.; Park, S.M. Precore and core promoter mutations of hepatitis B virus and hepatitis B e antigen-negative chronic hepatitis B in Korea. J. Hepatol. 2003, 38, 98–103. [Google Scholar] [CrossRef]
- Song, B.C.; Kim, H.; Kim, S.H.; Cha, C.Y.; Kook, Y.H.; Kim, B.J. Comparison of full length sequences of hepatitis B virus isolates in hepatocellular carcinoma patients and asymptomatic carriers of Korea. J. Med. Virol. 2005, 75, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Song, B.C.; Kim, S.H.; Kim, H.; Ying, Y.H.; Kim, H.J.; Kim, Y.J.; Yoon, J.H.; Lee, H.S.; Cha, C.Y.; Kook, Y.H.; et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J. Med. Virol. 2005, 76, 194–202. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jee, Y.; Mun, H.S.; Park, J.H.; Yoon, J.H.; Kim, Y.J.; Lee, H.S.; Hyun, J.W.; Hwang, E.S.; Cha, C.Y.; et al. Characterization of two hepatitis B virus populations in a single Korean hepatocellular carcinoma patient with an HBeAg-negative serostatus: A novel X-Gene-deleted strain with inverted duplication sequences of upstream enhancer site II. Intervirology 2007, 50, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jee, Y.; Mun, H.S.; Song, B.C.; Park, J.H.; Hyun, J.W.; Hwang, E.S.; Cha, C.Y.; Kook, Y.H.; Kim, B.J. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J. Microbiol. Biotechnol. 2007, 17, 701–704. [Google Scholar] [PubMed]
- Kim, H.J.; Park, J.H.; Jee, Y.; Lee, S.A.; Kim, H.; Song, B.C.; Yang, S.; Lee, M.; Yoon, J.H.; Kim, Y.J.; et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J. Med. Virol. 2008, 80, 1337–1343. [Google Scholar] [CrossRef] [PubMed]
- Mun, H.S.; Lee, S.A.; Jee, Y.; Kim, H.; Park, J.H.; Song, B.C.; Yoon, J.H.; Kim, Y.J.; Lee, H.S.; Hyun, J.W.; et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J. Med. Virol. 2008, 80, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Cho, Y.K.; Lee, K.H.; Hwang, E.S.; Kook, Y.H.; Kim, B.J. Gender disparity in distribution of the major hydrophilic region variants of hepatitis B virus genotype C according to hepatitis B e antigen serostatus. J. Med. Virol. 2011, 83, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Mun, H.S.; Kim, H.; Lee, H.K.; Kim, B.J.; Hwang, E.S.; Kook, Y.H.; Kim, B.J. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J. Med. Virol. 2011, 83, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Mun, H.S.; Lee, S.A.; Kim, H.; Hwang, E.S.; Kook, Y.H.; Kim, B.J. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J. Virol. 2011, 85, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, S.A.; Hwang, E.S.; Kook, Y.H.; Kim, B.J. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS ONE 2012, 7, e47372. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, K.; Kim, H.; Kim, B.J. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J. Hepatol. 2012, 56, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.A.; Kim, D.W.; Lee, S.H.; Kim, B.J. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS ONE 2013, 8, e54486. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, K.J.; Kim, D.W.; Kim, B.J. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J. Clin. Microbiol. 2013, 51, 3928–3936. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J. Gastroenterol. 2014, 20, 460–467. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.W.; Lee, S.A.; Kim, H.; Won, Y.S.; Kim, B.J. Naturally occurring mutations in the nonstructural region 5B of hepatitis C virus (HCV) from treatment-naive Korean patients chronically infected with HCV genotype 1b. PLoS ONE 2014, 9, e87773. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, H.; Won, Y.S.; Seok, S.H.; Na, Y.; Shin, H.B.; Inn, K.S.; Kim, B.J. Male-specific hepatitis B virus large surface protein variant W4P potentiates tumorigenicity and induces gender disparity. Mol. Cancer 2015, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, B.J. Association of preS/S Mutations with Occult Hepatitis B Virus (HBV) Infection in South Korea: Transmission Potential of Distinct Occult HBV Variants. Int. J. Mol. Sci. 2015, 16, 13595–13609. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.A.; Kim, B.J. X region mutations of hepatitis B virus related to clinical severity. World J. Gastroenterol. 2016, 22, 5467–5478. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, S.A.; Do, S.Y.; Kim, B.J. Precore/core region mutations of hepatitis B virus related to clinical severity. World J. Gastroenterol. 2016, 22, 4287–4296. [Google Scholar] [CrossRef] [PubMed]
- Sugauchi, F.; Orito, E.; Ichida, T.; Kato, H.; Sakugawa, H.; Kakumu, S.; Ishida, T.; Chutaputti, A.; Lai, C.L.; Ueda, R.; et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J. Virol. 2002, 76, 5985–5992. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Xing, K.; Deng, R.; Wang, J.; Wang, X. Identification of Hepatitis B virus putative intergenotype recombinants by using fragment typing. J. Gen. Virol. 2006, 87, 2203–2215. [Google Scholar] [CrossRef] [PubMed]
- Rehermann, B.; Nascimbeni, M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005, 5, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Kurbanov, F.; Tanaka, Y.; Fujiwara, K.; Sugauchi, F.; Mbanya, D.; Zekeng, L.; Ndembi, N.; Ngansop, C.; Kaptue, L.; Miura, T.; et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J. Gen. Virol. 2005, 86, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Mulyanto; Pancawardani, P.; Depamede, S.N.; Wahyono, A.; Jirintai, S.; Nagashima, S.; Takahashi, M.; Nishizawa, T.; Okamoto, H. Identification of four novel subgenotypes (C13-C16) and two inter-genotypic recombinants (C12/G and C13/B3) of hepatitis B virus in Papua province, Indonesia. Virus Res. 2012, 163, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Cao, G.W. Clinical relevance and public health significance of hepatitis B virus genomic variations. World J. Gastroenterol. 2009, 15, 5761–5769. [Google Scholar] [CrossRef] [PubMed]
- Fares, M.A.; Holmes, E.C. A revised evolutionary history of hepatitis B virus (HBV). J. Mol. Evol. 2002, 54, 807–814. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, C.; Krogsgaard, K.; Horal, P.; Lindh, M. Interpred Trial Group. Genotype mixtures of hepatitis B virus in patients treated with interferon. J. Infect. Dis. 2002, 186, 752–759. [Google Scholar] [CrossRef] [PubMed]
- Gerner, P.R.; Friedt, M.; Oettinger, R.; Lausch, E.; Wirth, S. The hepatitis B virus seroconversion to anti-HBe is frequently associated with HBV genotype changes and selection of preS2-defective particles in chronically infected children. Virology 1998, 245, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.; Orito, E.; Gish, R.G.; Sugauchi, F.; Suzuki, S.; Ueda, R.; Miyakawa, Y.; Mizokami, M. Characteristics of hepatitis B virus isolates of genotype G and their phylogenetic differences from the other six genotypes (A through F). J. Virol. 2002, 76, 6131–6137. [Google Scholar] [CrossRef] [PubMed]
- Morozov, V.; Pisareva, M.; Groudinin, M. Homologous recombination between different genotypes of hepatitis B virus. Gene 2000, 260, 55–65. [Google Scholar] [CrossRef]
- Cui, C.; Shi, J.; Hui, L.; Xi, H.; Zhuoma; Quni; Tsedan; Hu, G. The dominant hepatitis B virus genotype identified in Tibet is a C/D hybrid. J. Gen. Virol. 2002, 83, 2773–2777. [Google Scholar] [CrossRef] [PubMed]
- Kimbi, G.C.; Kramvis, A.; Kew, M.C. Distinctive sequence characteristics of subgenotype A1 isolates of hepatitis B virus from South Africa. J. Gen. Virol. 2004, 85, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.H.; Jeng, K.S.; Syu, W.J.; Huang, Y.H.; Su, C.W.; Peng, W.L.; Sheen, I.J.; Wu, J.C. Hepatitis B surface antigen levels and sequences of natural hepatitis B virus variants influence the assembly and secretion of hepatitis d virus. J. Virol. 2008, 82, 2250–2264. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lok, A.S. Viral factors and outcomes of chronic HBV infection. Am. J. Gastroenterol. 2011, 106, 93–95. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hou, J.; Zeng, G.; Wen, S.; Tanaka, Y.; Cheng, J.; Kurbanov, F.; Wang, L.; Jiang, J.; Naoumov, N.V.; Mizokami, M.; Qi, Y. Distribution and characteristics of hepatitis B virus genotype C subgenotypes in China. J. Viral Hepat 2007, 14, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Tanaka, Y.; Hige, S.; Yamada, G.; Murawaki, Y.; Komatsu, M.; Kuramitsu, T.; Kawata, S.; Tanaka, E.; Izumi, N.; et al. Distribution of hepatitis B virus genotypes among patients with chronic infection in Japan shifting toward an increase of genotype A. J. Clin. Microbiol. 2009, 47, 1476–1483. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence-Limits on Phylogenies—An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-Y.; Lee, S.-H.; Kim, J.-E.; Kim, H.; Kim, K.; Kook, Y.-H.; Kim, B.-J. Identification of Novel A2/C2 Inter-Genotype Recombinants of Hepatitis B Virus from a Korean Chronic Patient Co-Infected with Both Genotype A2 and C2. Int. J. Mol. Sci. 2017, 18, 737. https://doi.org/10.3390/ijms18040737
Lee S-Y, Lee S-H, Kim J-E, Kim H, Kim K, Kook Y-H, Kim B-J. Identification of Novel A2/C2 Inter-Genotype Recombinants of Hepatitis B Virus from a Korean Chronic Patient Co-Infected with Both Genotype A2 and C2. International Journal of Molecular Sciences. 2017; 18(4):737. https://doi.org/10.3390/ijms18040737
Chicago/Turabian StyleLee, So-Young, Seung-Hee Lee, Ji-Eun Kim, Hong Kim, Kijeong Kim, Yoon-Hoh Kook, and Bum-Joon Kim. 2017. "Identification of Novel A2/C2 Inter-Genotype Recombinants of Hepatitis B Virus from a Korean Chronic Patient Co-Infected with Both Genotype A2 and C2" International Journal of Molecular Sciences 18, no. 4: 737. https://doi.org/10.3390/ijms18040737





