Early Assessment of Colorectal Cancer by Quantifying Circulating Tumor Cells in Peripheral Blood: ECT2 in Diagnosis of Colorectal Cancer
Abstract
:1. Introduction
2. Results
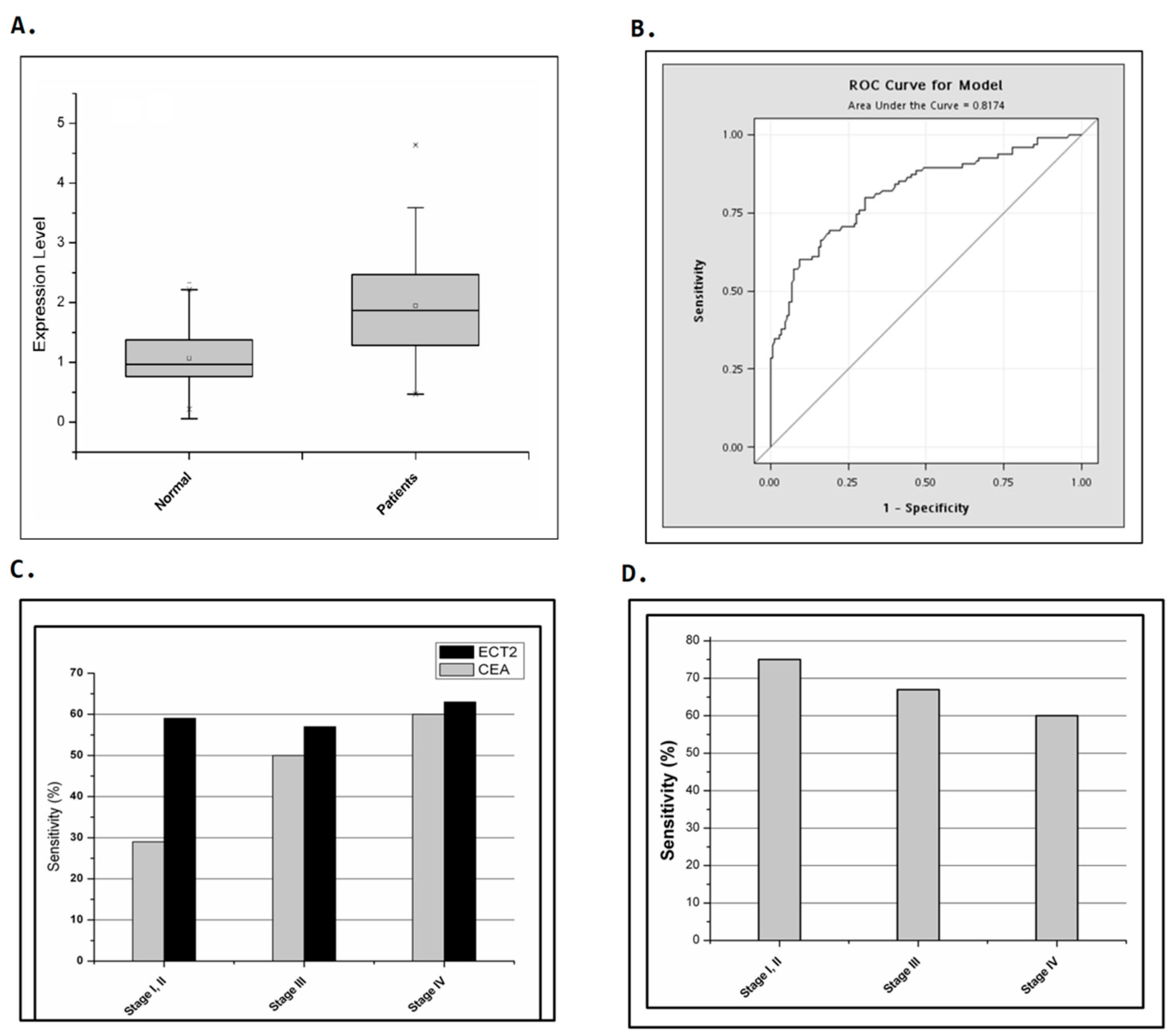
2.1. Identification of ECT2 as a Candidate Marker Gene for Quantifying Circulating Tumor Cells in Colorectal Cancer Patients
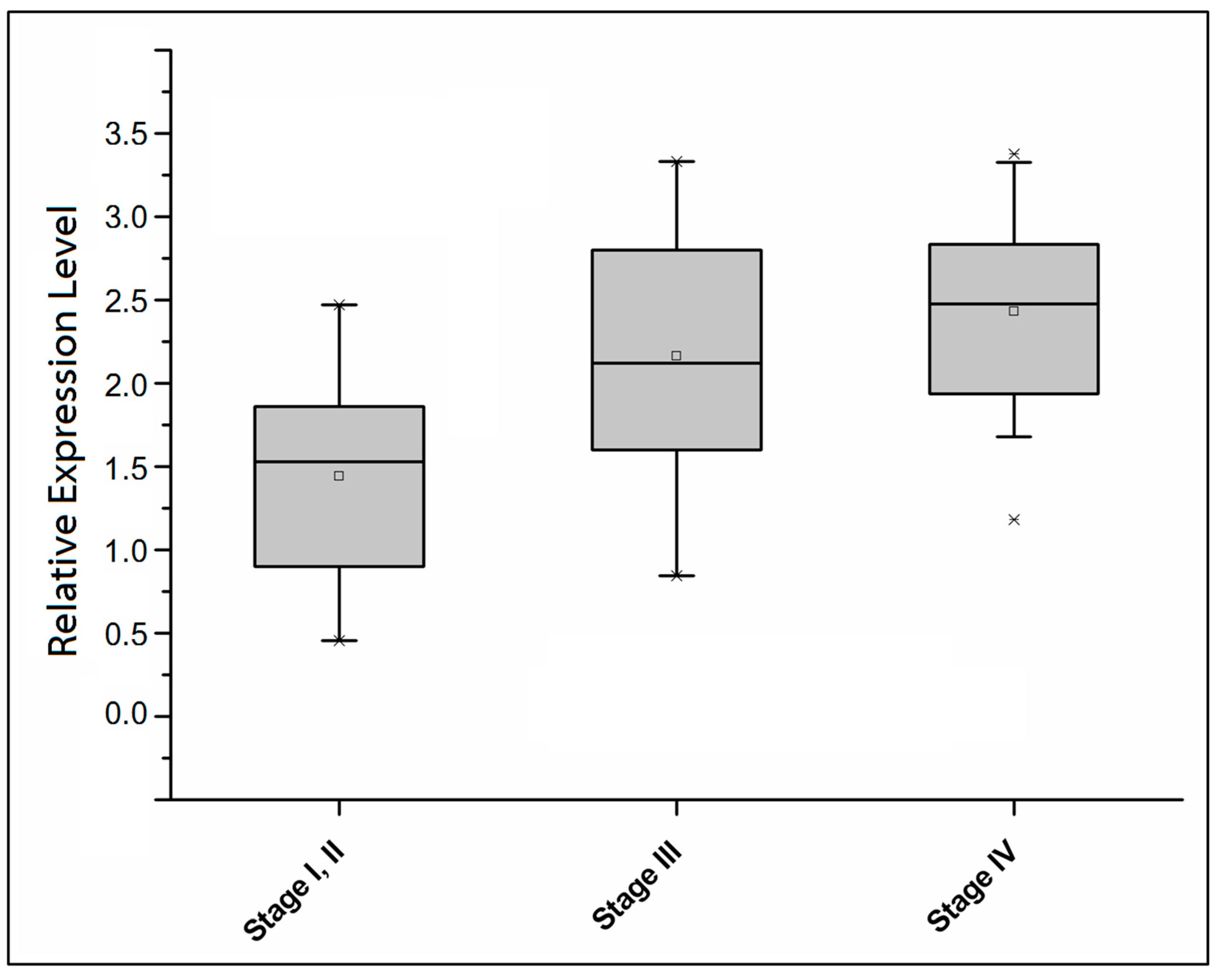
2.2. Comparison of the Detection Sensitivity of CEA and ECT2 in Colorectal Cancer Patients at Different Stages
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Study Subjects
4.3. Sample Collection and RNA Preparation of Circulating Tumor Cells
4.4. Microarray Analysis and Nested Real-Time Quantitative Polymerase Chain Reactions
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CTCs | Circulating tumor cells |
| ECT2 | Epithelial cell transforming sequence 2 oncogene |
| AUC | Area under the curve |
| CEA | Carcinoembryonic antigen |
| EMT | Epithelial to mesenchymal transition |
| qPCR | Quantitative PCR |
| KW | Kruskal-Wallis |
References
- Weiser, M.R.; Gonen, M.; Chou, J.F.; Kattan, M.W.; Schrag, D. Predicting survival after curative colectomy for cancer: Individualizing colon cancer staging. J. Clin. Oncol. 2011, 29, 4796–4802. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Cancer survivors—United States, 2007. MMWR Morb. Mortal. Wkly. Rep. 2011, 60, 269–272. [Google Scholar]
- Siegel, R.; DeSantis, C.; Virgo, K.; Stein, K.; Mariotto, A.; Smith, T.; Cooper, D.; Gansler, T.; Lerro, C.; Fedewa, S.; et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 2012, 62, 220–241. [Google Scholar] [CrossRef] [PubMed]
- Ku, G.; Tan, I.B.; Yau, T.; Boku, N.; Laohavinij, S.; Cheng, A.L.; Kang, Y.K.; de Lima Lopes, G., Jr. Management of colon cancer: Resource-stratified guidelines from the Asian Oncology Summit 2012. Lancet Oncol. 2012, 13, e470–e481. [Google Scholar] [CrossRef]
- Sharma, M.R.; Schilsky, R.L. GI cancers in 2010: New standards and a predictive biomarker for adjuvant therapy. Nat. Rev. Clin. Oncol. 2011, 8, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Galandiuk, S. Induction chemoradiation for rectal cancer. Arch. Surg. 2006, 141, 1246–1252; discussion 1253. [Google Scholar] [CrossRef] [PubMed]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; Gorges, T.M.; Pantel, K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 2015, 7, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Lim, C.T. Tumor dissemination: An EMT affair. Cancer Cell 2013, 23, 272–273. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Heller, G.; Molina, A.; Attard, G.; Danila, D.C.; Jia, X.; Peng, W.; Sandhu, S.K.; Olmos, D.; Riisnaes, R.; et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2015, 33, 1348–1355. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Mitra, A.; Brownlee, Z.; Xia, X.; Bellister, S.; Overman, M.J.; Kopetz, S.; Ellis, L.M.; Meng, Q.H.; Li, S. Epithelial-mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin. Cancer Res. 2015, 21, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef] [PubMed]
- Yokobori, T.; Iinuma, H.; Shimamura, T.; Imoto, S.; Sugimachi, K.; Ishii, H.; Iwatsuki, M.; Ota, D.; Ohkuma, M.; Iwaya, T.; et al. Plastin3 is a novel marker for circulating tumor cells undergoing the epithelial-mesenchymal transition and is associated with colorectal cancer prognosis. Cancer Res. 2013, 73, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Tang, R.N.; Zou, J.H.; Wang, F.C. The prognostic role of disseminated tumor cells detected in peripheral blood and bone marrow of colorectal cancer. Hepato Gastroenterol. 2012, 59, 2164–2167. [Google Scholar]
- Vardakis, N.; Messaritakis, I.; Papadaki, C.; Agoglossakis, G.; Sfakianaki, M.; Saridaki, Z.; Apostolaki, S.; Koutroubakis, I.; Perraki, M.; Hatzidaki, D.; et al. Prognostic significance of the detection of peripheral blood CEACAM5mRNA-positive cells by real-time polymerase chain reaction in operable colorectal cancer. Clin. Cancer Res. 2011, 17, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; van Dalen, A.; Haglund, C.; Hansson, L.; Holinski-Feder, E.; Klapdor, R.; Lamerz, R.; Peltomaki, P.; Sturgeon, C.; Topolcan, O. Tumour markers in colorectal cancer: European Group on Tumour Markers (EGTM) guidelines for clinical use. Eur. J. Cancer 2007, 43, 1348–1360. [Google Scholar] [CrossRef] [PubMed]
- Bacolod, M.D.; Barany, F. Molecular profiling of colon tumors: The search for clinically relevant biomarkers of progression, prognosis, therapeutics, and predisposition. Ann. Surg. Oncol. 2011, 18, 3694–3700. [Google Scholar] [CrossRef] [PubMed]
- Hampton, R.; Walker, M.; Marshall, J.; Juhl, H. Differential expression of carcinoembryonic antigen (CEA) splice variants in whole blood of colon cancer patients and healthy volunteers: Implication for the detection of circulating colon cancer cells. Oncogene 2002, 21, 7817–7823. [Google Scholar] [CrossRef] [PubMed]
- Bolocan, A.; Ion, D.; Ciocan, D.N.; Paduraru, D.N. Prognostic and predictive factors in colorectal cancer. Chirurgia 2012, 107, 555–563. [Google Scholar] [PubMed]
- Fakih, M.G.; Padmanabhan, A. CEA monitoring in colorectal cancer. What you should know. Oncology 2006, 20, 579–587; discussion 588, 594, 596 passim. [Google Scholar] [PubMed]
- Lu, C.Y.; Tsai, H.L.; Uen, Y.H.; Hu, H.M.; Chen, C.W.; Cheng, T.L.; Lin, S.R.; Wang, J.Y. Circulating tumor cells as a surrogate marker for determining clinical outcome to mFOLFOX chemotherapy in patients with stage III colon cancer. Br. J. Cancer 2013, 108, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabieres, C.; Schwarzenbach, H.; Pantel, K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Molnar, B.; Ladanyi, A.; Tanko, L.; Sreter, L.; Tulassay, Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clin. Cancer Res. 2001, 7, 4080–4085. [Google Scholar] [PubMed]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; Miller, M.C.; et al. Prognostic significance of circulating tumor cells in patients with metastatic colorectal cancer. Ann. Oncol. 2009, 20, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Tol, J.; Koopman, M.; Miller, M.C.; Tibbe, A.; Cats, A.; Creemers, G.J.; Vos, A.H.; Nagtegaal, I.D.; Terstappen, L.W.; Punt, C.J. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann. Oncol. 2010, 21, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Coumans, F.A.; Doggen, C.J.; Attard, G.; de Bono, J.S.; Terstappen, L.W. All circulating EpCAM+CK+CD45− objects predict overall survival in castration-resistant prostate cancer. Ann. Oncol. 2010, 21, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Smith, C.L.; Long, J.E.; Eva, A.; Fleming, T.P. Oncogene ECT2 is related to regulators of small GTP-binding proteins. Nature 1993, 362, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Tatsumoto, T.; Xie, X.; Blumenthal, R.; Okamoto, I.; Miki, T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J. Cell Biol. 1999, 147, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Hara, T.; Abe, M.; Inoue, H.; Yu, L.R.; Veenstra, T.D.; Kang, Y.H.; Lee, K.S.; Miki, T. Cytokinesis regulator ECT2 changes its conformation through phosphorylation at Thr-341 in G2/M phase. Oncogene 2006, 25, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Miki, T. Interaction of ect2 and Dbl with Rho-related GTPases. Methods Enzymol. 1995, 256, 90–98. [Google Scholar] [PubMed]
- Kim, J.E.; Billadeau, D.D.; Chen, J. The tandem BRCT domains of ECT2 are required for both negative and positive regulation of ECT2 in cytokinesis. J. Biol. Chem. 2005, 280, 5733–5739. [Google Scholar] [CrossRef] [PubMed]
- Hirata, D.; Yamabuki, T.; Miki, D.; Ito, T.; Tsuchiya, E.; Fujita, M.; Hosokawa, M.; Chayama, K.; Nakamura, Y.; Daigo, Y. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin. Cancer Res. 2009, 15, 256–266. [Google Scholar] [CrossRef] [PubMed]
- Wondergem, B.; Zhang, Z.; Huang, D.; Ong, C.K.; Koeman, J.; Hof, D.V.; Petillo, D.; Ooi, A.; Anema, J.; Lane, B.; et al. Expression of the PTTG1 oncogene is associated with aggressive clear cell renal cell carcinoma. Cancer Res. 2012, 72, 4361–4371. [Google Scholar] [CrossRef] [PubMed]
- Fortin, S.P.; Ennis, M.J.; Schumacher, C.A.; Zylstra-Diegel, C.R.; Williams, B.O.; Ross, J.T.; Winkles, J.A.; Loftus, J.C.; Symons, M.H.; Tran, N.L. Cdc42 and the guanine nucleotide exchange factors ECT2 and trio mediate Fn14-induced migration and invasion of glioblastoma cells. Mol. Cancer Res. 2012, 10, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.; Lee, S.; Choi, H.S.; Kim, S.N.; Lee, E.; Shin, Y.; Seo, J.; Kim, B.; Kim, W.K.; Chun, H.K.; Lee, W.Y.; Kim, J. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin. Cancer Res. 2011, 17, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; Pantel, K. Biologic challenges in the detection of circulating tumor cells. Cancer Res. 2013, 73, 8–11. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.S.; Kerr, M.A. Lewis blood group and CEA related antigens; coexpressed cell-cell adhesion molecules with roles in the biological progression and dissemination of tumours. Mol. Pathol. 1999, 52, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Peck, K.; Sher, Y.P.; Shih, J.Y.; Roffler, S.R.; Wu, C.W.; Yang, P.C. Detection and quantitation of circulating cancer cells in the peripheral blood of lung cancer patients. Cancer Res. 1998, 58, 2761–2765. [Google Scholar] [PubMed]
- Sher, Y.P.; Shih, J.Y.; Yang, P.C.; Roffler, S.R.; Chu, Y.W.; Wu, C.W.; Yu, C.L.; Peck, K. Prognosis of non-small cell lung cancer patients by detecting circulating cancer cells in the peripheral blood with multiple marker genes. Clin. Cancer Res. 2005, 11, 173–179. [Google Scholar] [PubMed]

| Patients (n = 90) | Control (n = 151) | |||
|---|---|---|---|---|
| Characteristic | No. | % | No. | % |
| Age, year | ||||
| Mean | 66.45 | 36.29 | ||
| Range | 36–90 | 22–69 | ||
| Gender | ||||
| Female | 45 | 50 | 101 | 67 |
| Male | 45 | 50 | 50 | 33 |
| Primary Tumor | ||||
| Colon | 59 | 61 | N/A | |
| Rectal | 33 | 34 | ||
| Rectosigmoid | 3 | 3 | ||
| Others | 2 | 2 | ||
| Stage | ||||
| I | 6 | 6 | N/A | |
| II | 11 | 12 | ||
| III | 30 | 33 | ||
| IV | 43 | 47 | ||
| Differentiation | ||||
| Well | 2 | 2 | N/A | |
| Moderate | 81 | 90 | ||
| Poor | 5 | 6 | ||
| Unspecified | 2 | 2 | ||
| CEA, ng/mL | ||||
| ≥5 | 46 | 51 | N/A | |
| <5 | 44 | 49 | ||
| CEA/ECT Expression | Stage I and II, n = 17 (%) | Stage III, n = 30 (%) | Stage IV, n = 43 (%) | All, n = 90 (%) |
|---|---|---|---|---|
| CEA+ECT2− | 4 (24) | 9 (30) | 9 (21) | 22 (24) |
| CEA−ECT2+ | 9 (53) | 11 (37) | 10 (23) | 30 (33) |
| CEA+ECT2+ | 1 (6) | 6 (20) | 6 (20) | 24 (27) |
| CEA+ and/or ECT2+ | 14 (82) | 26 (87) | 26 (84) | 76 (84) |
| CEA−ECT2− | 3 (18) | 4 (13) | 7 (16) | 14 (16) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-J.; Sung, W.-W.; Chen, H.-C.; Chern, Y.-J.; Hsu, H.-T.; Lin, Y.-M.; Lin, S.-H.; Peck, K.; Yeh, K.-T. Early Assessment of Colorectal Cancer by Quantifying Circulating Tumor Cells in Peripheral Blood: ECT2 in Diagnosis of Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 743. https://doi.org/10.3390/ijms18040743
Chen C-J, Sung W-W, Chen H-C, Chern Y-J, Hsu H-T, Lin Y-M, Lin S-H, Peck K, Yeh K-T. Early Assessment of Colorectal Cancer by Quantifying Circulating Tumor Cells in Peripheral Blood: ECT2 in Diagnosis of Colorectal Cancer. International Journal of Molecular Sciences. 2017; 18(4):743. https://doi.org/10.3390/ijms18040743
Chicago/Turabian StyleChen, Chih-Jung, Wen-Wei Sung, Hung-Chang Chen, Yi-Jye Chern, Hui-Ting Hsu, Yueh-Min Lin, Shu-Hui Lin, Konan Peck, and Kun-Tu Yeh. 2017. "Early Assessment of Colorectal Cancer by Quantifying Circulating Tumor Cells in Peripheral Blood: ECT2 in Diagnosis of Colorectal Cancer" International Journal of Molecular Sciences 18, no. 4: 743. https://doi.org/10.3390/ijms18040743





