Sus scrofa miR-204 and miR-4331 Negatively Regulate Swine H1N1/2009 Influenza A Virus Replication by Targeting Viral HA and NS, Respectively
Abstract
:1. Introduction
2. Results
2.1. Prediction of Potential Swine miRNAs Targeting SIV-H1N1/2009 Genomic RNA
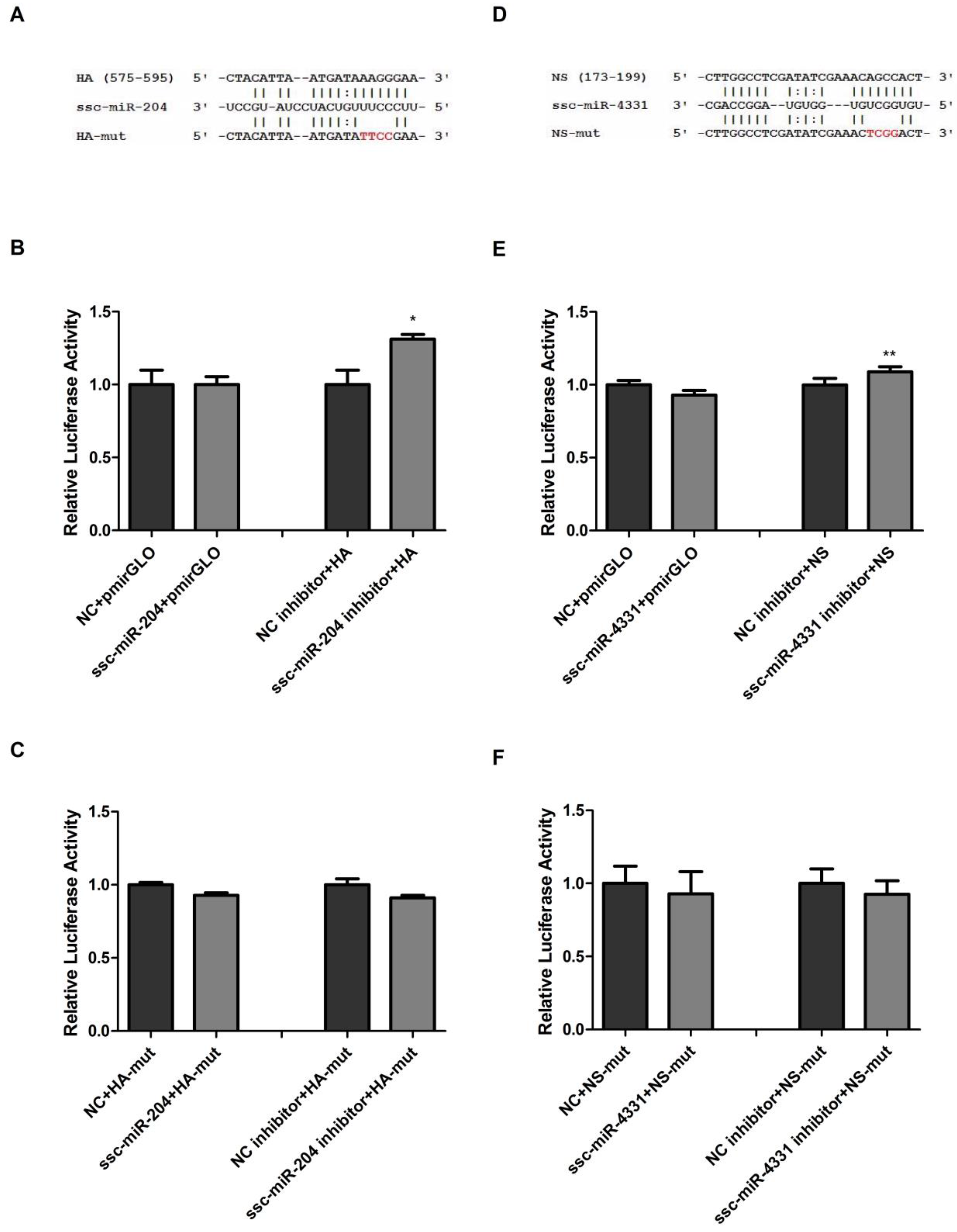
2.2. Validation of ssc-miR-204 and ssc-miR-4331 Targeting Viral HA and NS of SIV-H1N1/2009, Respectively
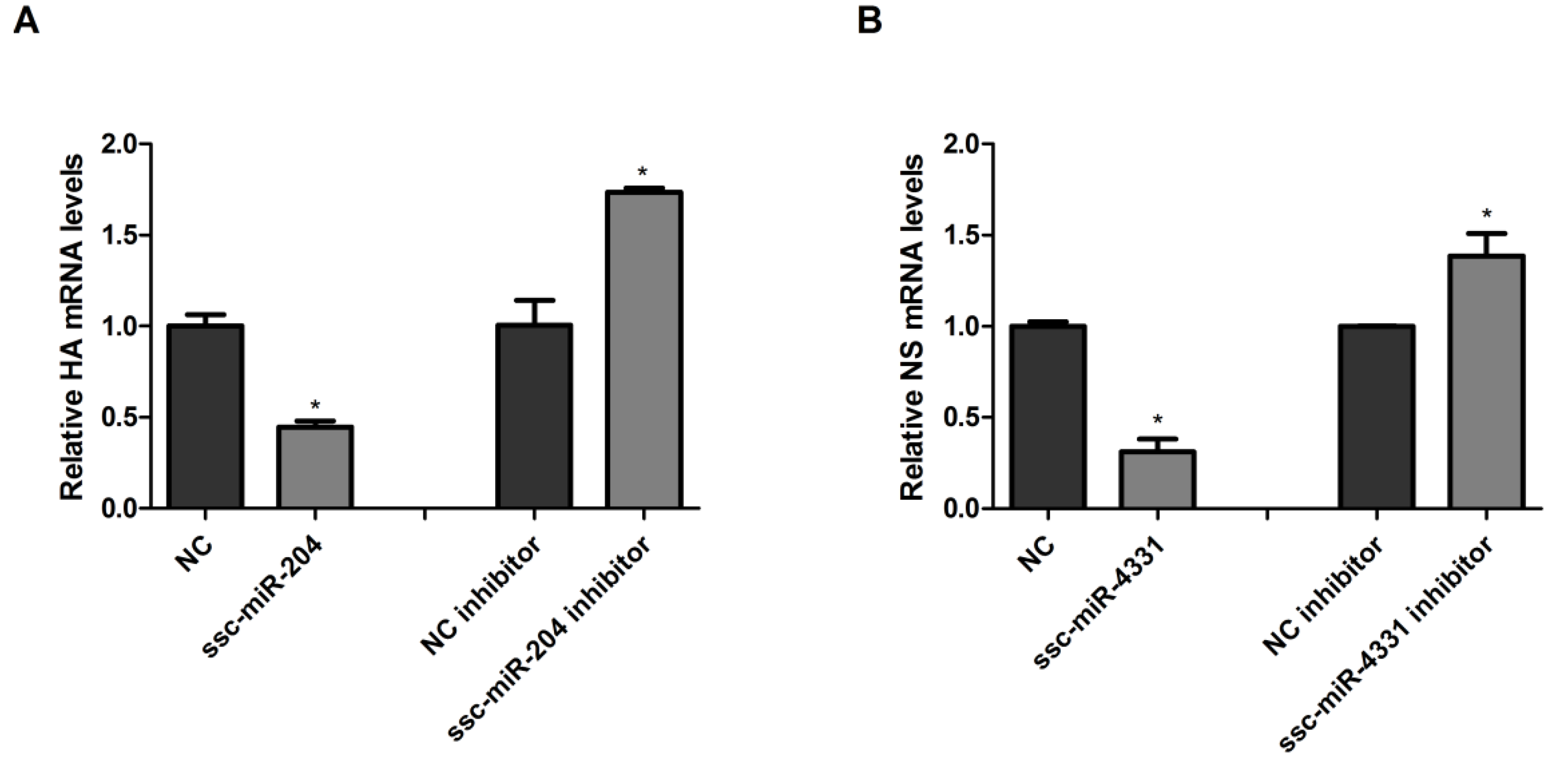
2.3. Ssc-miR-204 and ssc-miR-4331 Repressed the Expression of Viral HA1 and NS1 Respectively, and Inhibited the Replication of SIV-H1N1/2009
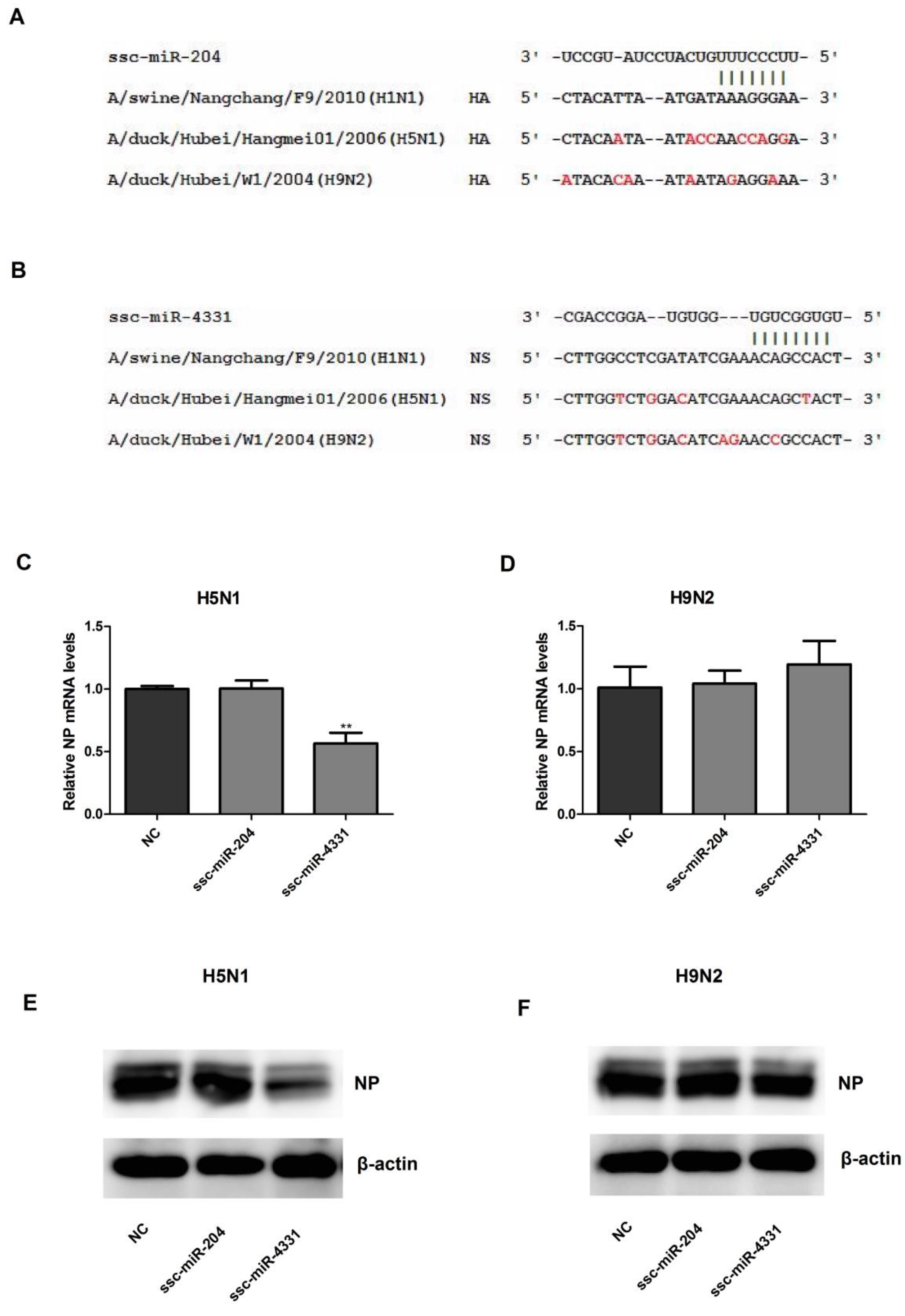
2.4. The Effect of ssc-miR-204 and ssc-miR-4331 on the Replication of H5N1 or H9N2 Influenza A Virus
2.5. SIV-H1N1/2009 Infection Downregulated the Expression of ssc-miR-204 and ssc-miR-4331
3. Discussion
4. Materials and Methods
4.1. Cells Culture
4.2. Virus Infection
4.3. Plasmid Construction
4.4. Ssc-miR-204 and ssc-miR-4331 Mimics and Inhibitor
4.5. Transfection
4.6. Potential miRNAs Prediction and Dual-Luciferase Reporter Assays
4.7. RNA Extraction, Reverse Transcription and qRT-PCR
4.8. Western Blotting
4.9. Neuraminidase Activity Measured by a MUNANA Assay
4.10. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Palese, P.; Shaw, M.L. Orthomyxoviridae: The viruses and their replication. Fields Virol. 2007, 2, 1647–1689. [Google Scholar]
- Steinhauer, D.A.; Skehel, J.J. Genetics of influenza viruses. Annu. Rev. Genet. 2002, 36, 305–332. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Calvo, P.A.; Malide, D.; Gibbs, J.; Schubert, U.; Bacik, I.; Basta, S.; O’Neill, R.; Schickli, J.; Palese, P.; et al. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 2001, 7, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.M.; Barbezange, C.; Jagger, B.W.; Dalton, R.M.; Gog, J.R.; Curran, M.D.; Taubenberger, J.K.; Anderson, E.C.; Digard, P. Overlapping signals for translational regulation and packaging of influenza A virus segment 2. Nucleic Acids Res. 2011, 39, 7775–7790. [Google Scholar] [CrossRef] [PubMed]
- Jagger, B.W.; Wise, H.M.; Kash, J.C.; Walters, K.A.; Wills, N.M.; Xiao, Y.L.; Dunfee, R.L.; Schwartzman, L.M.; Ozinsky, A.; Bell, G.L.; et al. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 2012, 337, 199–204. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, Y.; Noda, T.; Kawakami, E.; Akkina, R.; Kawaoka, Y. Identification of novel influenza A virus proteins translated from PA mRNA. J. Virol. 2012, 87, 2455–2462. [Google Scholar] [CrossRef] [PubMed]
- Wise, H.M.; Hutchinson, E.C.; Jagger, B.W.; Stuart, A.D.; Kang, Z.H.; Robb, N.; Schwartzman, L.M.; Kash, J.C.; Fodor, E.; Firth, A.E.; et al. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012, 8, e1002998. [Google Scholar] [CrossRef] [PubMed]
- Selman, M.; Dankar, S.K.; Forbes, N.E.; Jia, J.J.; Brown, E.G. Adaptive mutation in influenza A virus non-structural gene is linked to host switching and induces a novel protein by alternative splicing. Emerg. Microbes Infect. 2012, 1, e42. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wu, Y.; Tefsen, B.; Shi, Y.; Gao, G.F. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014, 22, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Matrosovich, M.; Stech, J.; Klenk, H.D. Influenza receptors, polymerase and host range. Rev. Sci. Tech. OIE 2009, 28, 203–217. [Google Scholar] [CrossRef]
- Smith, G.J.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, S.U.; Schnitzler, P. An update on swine-origin influenza virus A/H1N1: A review. Virus Genes 2009, 39, 279. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Jain, S.; Finelli, L.; Shaw, M.W.; Lindstrom, S.; Garten, R.J.; Gubareva, R.J.; Xu, X.; Bridges, C.B.; Uyeki, T.M. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 2009, 2009, 2605–2615. [Google Scholar]
- World Health Organization. Pandemic (H1N1) 2009-Update 112; 2010; Available online: http://www.who.int/csr/don/2010_08_06/en/ (accessed on 29 March 2017).
- Howden, K.J.; Brockhoff, E.J.; Caya, F.D.; McLeod, L.J.; Lavoie, M.; Ing, J.D.; Bystrom, J.M.; Alexandersen, S.; Pasick, J.M.; Berhane, Y.; et al. An investigation into human pandemic influenza virus (H1N1) 2009 on an Alberta swine farm. Can. Vet. J. 2009, 50, 1153–1161. [Google Scholar] [PubMed]
- Chen, H.; Wang, Y.; Liu, W.; Zhang, J.; Dong, B.; Fan, X.; de Jong, M.D.; Farrar, J.; Riley, S.; Smith, G.J.D.; et al. Serologic survey of pandemic (H1N1) 2009 virus, Guangxi Province, China. Emerg. Infect. Dis. 2009, 15, 1849–1850. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Wang, C.; Yang, Y.; Guo, X.; Kang, C.; Chen, H.; Jin, M. Pandemic (H1N1) 2009 virus in swine herds, People’s Republic of China. Emerg. Infect. Dis. 2011, 17, 1757–1759. [Google Scholar] [CrossRef] [PubMed]
- Sreta, D.; Tantawet, S.; Na Ayudhya, S.N.; Thontiravong, A.; Wongphatcharachai, M.; Lapkuntod, J.; Bunpapong, N.; Tuanudom, R.; Suradhat, S.; Vimolket, L.; et al. Pandemic (H1N1) 2009 virus on commercial swine farm, Thailand. Emerg. Infect. Dis. 2010, 16, 1587–1591. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Lee, J.H.; Pascua, P.N.; Baek, Y.H.; Kwon, H.I.; Park, K.J.; Choi, H.W.; Shin, Y.K.; Song, J.Y.; Kim, C.J.; et al. Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J. Clin. Microbiol. 2010, 48, 3204–3211. [Google Scholar] [CrossRef] [PubMed]
- Howard, W.A.; Essen, S.C.; Strugnell, B.W.; Russell, C.; Barass, L.; Reid, S.M.; Brown, I.H. Reassortant Pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg. Infect. Dis. 2011, 17, 1049–1052. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Couceiro, J.N.S.; Kelm, S.; Baum, L.G.; Krauss, S.; Castrucci, M.R.; Donatelli, I.; Kida, H.; Paulson, J.C.; Webster, R.G.; et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998, 72, 7367–7373. [Google Scholar] [PubMed]
- Vijaykrishna, D.; Poon, L.L.; Zhu, H.C.; Ma, S.K.; Li, O.T.; Cheung, C.L.; Smith, G.J.; Peiris, J.S.; Guan, Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 2010, 328, 1529. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Di Trani, L.; Faccini, S.; Vaccari, G.; Nigrelli, D.; Boniotti, M.B.; Falcone, E.; Boni, A.; Chiapponi, C.; Sozzi, E.; et al. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet. Microbiol. 2010, 149, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Zhou, B.; Fan, X.; Lam, T.T.; Wang, J.; Chen, A.; Chen, X.; Chen, H.; Webster, R.G.; Webby, R.; et al. Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: Infectious potential for humans. J. Virol. 2011, 85, 10432–10439. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.J.; Pan, Q.; Li, D.G.; Sun, H.; Liu, B.W. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J. Hepatol. 2009, 50, 766–778. [Google Scholar] [CrossRef] [PubMed]
- Scaria, V.; Hariharan, M.; Maiti, S.; Pillai, B.; Brahmachari, S.K. Host-virus interaction: A new role for microRNAs. Retrovirology 2006, 3, 68. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Noti, J.D.; Beezhold, D.H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014, 468, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Jiang, L.; Lai, W.; Qin, Y.; Zhang, T.; Wang, S.; Ye, X. MicroRNA-33a disturbs influenza A virus replication by targeting ARCN1 and inhibiting viral ribonucleoprotein activity. J. Gen. Virol. 2016, 97, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhu, J.; Zhou, H.; Zhao, Z.; Zou, Z.; Liu, X.; Lin, X.; Zhang, X.; Deng, X.; Wang, R.; et al. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci. Rep. 2015, 5, 14991. [Google Scholar] [CrossRef] [PubMed]
- Ingle, H.; Kumar, S.; Raut, A.A.; Mishra, A.; Kulkarni, D.D.; Kameyama, T.; Takaoka, A.; Akira, S.; Kumar, H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015, 8, ra126. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, H.; Gao, S.; Jiang, W.; Huang, W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010, 84, 8849–8860. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.J.; Yang, J.; Fan, X.L.; Zhao, H.B.; Hu, W.; Li, Z.P.; Yu, G.C.; Ding, X.R.; Wang, J.Z.; Bo, X.C.; et al. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 2012, 16, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Khongnomnan, K.; Makkoch, J.; Poomipak, W.; Poovorawan, Y.; Payungporn, S. Human miR-3145 inhibits influenza A viruses replication by targeting and silencing viral PB1 gene. Exp. Biol. Med. 2015, 240, 1630–1639. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zou, W.; Jia, G.; Ke, J.; Zhu, J.; Lin, X.; Zhou, H.; Jin, M. The 2009 pandemic (H1N1) viruses isolated from pigs show enhanced pathogenicity in mice. Vet. Res. 2013, 44, 41. [Google Scholar] [CrossRef] [PubMed]
- Patient, A. Swine Influenza A (H1N1) Infection in Two Children—Southern California, March–April 2009. MMWR 2009, 58, 400. [Google Scholar]
- He, T.; Feng, G.; Chen, H.; Wang, L.; Wang, Y. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation 2009, 4, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef] [PubMed]
- Hale, B.G.; Randall, R.E.; Ortín, J.; Jackson, D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89, 2359–2376. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.M.; Soibam, B.; Benham, A.L.; Gunaratne, P.; Liu, H.; Trakooljul, N.; Ing, N.; et al. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genom. 2012, 13, 278. [Google Scholar] [CrossRef] [PubMed]
- Tambyah, P.A.; Sepramaniam, S.; Ali, J.M.; Chai, S.C.; Swaminathan, P.; Armugam, A.; Jeyaseelan, K. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS ONE 2013, 8, e76811. [Google Scholar]
- Sharbati, S.; Friedländer, M.R.; Sharbati, J.; Hoeke, L.; Chen, W.; Keller, A.; Stahler, P.F.; Rajewsky, N.; Einspanier, R. Deciphering the porcine intestinal microRNA transcriptome. BMC Genom. 2010, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Pawlina, K.; Gurgul, A.; Oczkowicz, M.; Bugno-Poniewierska, M. The characteristics of the porcine (Sus. scrofa) liver miRNAome with the use of next generation sequencing. J. Appl. Genet. 2015, 56, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xi, Q.Y.; Ye, R.S.; Cheng, X.; Qi, Q.E.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; Jiang, Q.Y.; et al. Exploration of microRNAs in porcine milk exosomes. BMC Genom. 2014, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhao, X.; Huang, Y.; Xiang, H.; Zhang, W.; Tong, D. Transmissible Gastroenteritis Virus (TGEV) infection alters the expression of cellular microRNA species that affect transcription of TGEV Gene 7. Int. J. Biol. Sci. 2015, 11, 913. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, M.; Scalvini, A.; Losio, M.N.; Corradi, A.; Soncini, M.; Bignotti, E.; Milanesi, E.; Ajmone-Marsan, P.; Barlati, S.; Tonelli, M. Establishment and characterization of two new pig cell lines for use in virological diagnostic laboratories. J. Virol. Methods 2003, 107, 205–212. [Google Scholar] [CrossRef]
- Zou, W.; Yu, Z.; Zhou, H.; Tu, J.; Jin, M. Genetic characterization of an H5N1 avian influenza virus with neurovirulence in ducks. Virus Genes 2009, 38, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.J.; Xu, G.Y.; Zhou, H.B.; Yu, Z.J.; Zhang, A.D.; Song, Y.F.; Jin, M.L.; Chen, H.C. Evolutionary characterization of influenza virus A/duck/Hubei/W1/2004 (H9N2) isolated from central China. Virus Genes 2008, 36, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Tafforeau, L.; Chantier, T.; Pradezynski, F.; Pellet, J.; Mangeot, P.E.; Vidalain, P.O.; Andre, P.; Rabourdin-Combe, C.; Lotteau, V. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J. Virol. 2011, 85, 13010–13018. [Google Scholar] [CrossRef] [PubMed]



| Genomic RNA | Predicted miRNAs | Free-Energy | Score |
|---|---|---|---|
| PB1 | ssc-miR-107 | −22.77 | 155.00 |
| ssc-miR-504 | −20.21 | 158.00 | |
| PB2 | ssc-miR-27a | −22.26 | 151.00 |
| ssc-miR-181d-5p | −21.26 | 159.00 | |
| ssc-miR-664-3p | −22.06 | 156.00 | |
| ssc-miR-769-3p | −20.39 | 164.00 | |
| ssc-miR-361-3p | −27.73 | 157.00 | |
| ssc-miR-4334-5p | −22.00 | 153.00 | |
| PA | ssc-miR-146a-3p | −20.01 | 156.00 |
| ssc-miR-432-5p | −23.28 | 159.00 | |
| HA | ssc-miR-204 | −22.23 | 163.00 |
| ssc-miR-365-5p | −20.89 | 152.00 | |
| ssc-miR-361-3p | −25.01 | 152.00 | |
| ssc-miR-339-5p | −24.67 | 152.00 | |
| ssc-miR-4334-3p | −24.13 | 152.00 | |
| NA | ssc-miR-136 | −26.18 | 173.00 |
| NP | ssc-miR-136 | −24.75 | 180.00 |
| ssc-miR-205 | −20.05 | 155.00 | |
| ssc-miR-204 | −22.71 | 164.00 | |
| ssc-miR-769-3p | −22.61 | 152.00 | |
| NS | ssc-miR-1307 | −29.54 | 160.00 |
| ssc-miR-4331 | −28.03 | 164.00 |
| Genomic RNA | Predicted miRNAs | Free-Energy | Score |
|---|---|---|---|
| PB1 | ssc-miR-186 | −23.53 | 157.00 |
| ssc-miR-196a | −20.32 | 154.00 | |
| ssc-miR-133b | −23.79 | 160.00 | |
| ssc-miR-196b-5p | −22.05 | 158.00 | |
| ssc-miR-339-5p | −21.79 | 153.00 | |
| ssc-miR-4338 | −21.74 | 172.00 | |
| ssc-miR-129b | −20.61 | 168.00 | |
| PB2 | ssc-miR-27b | −22.47 | 159.00 |
| ssc-miR-664-3p | −23.23 | 153.00 | |
| ssc-miR-92a | −30.34 | 164.00 | |
| ssc-miR-92b-3p | −25.65 | 156.00 | |
| ssc-miR-1307 | −21.08 | 150.00 | |
| ssc-miR-758 | −25.20 | 152.00 | |
| ssc-miR-194-3p | −29.20 | 167.00 | |
| ssc-miR-652 | −20.41 | 150.00 | |
| ssc-miR-4334-5p | −22.69 | 153.00 | |
| ssc-miR-1271 | −21.95 | 153.00 | |
| ssc-miR-1296 | −22.36 | 151.00 | |
| PA | ssc-miR-219 | −22.93 | 166.00 |
| HA | ssc-miR-151-5p | −25.20 | 159.00 |
| ssc-miR-935 | −25.96 | 168.00 | |
| NA | ssc-miR-338 | −21.40 | 162.00 |
| ssc-miR-218 | −26.37 | 165.00 | |
| NP | ssc-miR-145 | −22.84 | 160.00 |
| ssc-miR-205 | −20.68 | 151.00 | |
| ssc-miR-205 | −26.16 | 150.00 | |
| ssc-miR-124a | −23.21 | 151.00 | |
| ssc-miR-30a-3p | −22.14 | 157.00 | |
| ssc-miR-487b | −22.78 | 152.00 | |
| ssc-miR-219 | −20.93 | 158.00 | |
| ssc-miR-1343 | −22.47 | 155.00 | |
| NS | ssc-miR-181a | −21.82 | 154.00 |
| ssc-miR-365-5p | −24.76 | 155.00 | |
| ssc-miR-1307 | −29.86 | 164.00 | |
| M | ssc-miR-4336 | −20.86 | 153.00 |
| ssc-miR-129b | −23.31 | 157.00 |
| Primers | Sequence (5′–3′) |
|---|---|
| pmirGLO-HA-F | CTAGCTAGCAGCAAAAGCAGGGGAAAAC |
| pmirGLO-HA-R | CCGCTCGAGTAGTAGAAACAAGGGTGTTTTTTTC |
| pmirGLO-HA-mut-F | ATTAATGATATTCCGAAAGAAGTCC |
| pmirGLO-HA-mut-R | GGACTTCTTTCGGAATATCATTAAT |
| pmirGLO-NS-F | TACGAGCTCGCAAAAGCAGGGTGACAA |
| pmirGLO-NS-R | TGCTCTAGATAGTAGAAACAAGGGTGTTTTTTAT |
| pmirGLO-NS-mut-F | GATATCGAAACTCGGACTCTTGTTG |
| pmirGLO-NS-mut-R | CAACAAGAGTCCGAGTTTCGATATC |
| pmirGLO-PB1-F | TACGAGCTCAGCAAAAGCAGGCAAACC |
| pmirGLO-PB1-R | CCGCTCGAGTAGTAGAAACAAGGCATTTTTTCA |
| pmirGLO-PB2-F | CCGCTCGAGAGCAAAAGCAGGTCAAATATATT |
| pmirGLO-PB2-R | TGCTCTAGATAGTAGAAACAAGGTCGTTTTAAAC |
| pmirGLO-PA-F | TACGAGCTCAGCAAAAGCAGGTACTGATCC |
| pmirGLO-PA -R | CCGCTCGAGTAGTAGAAACAAGGTACTTTTTTGG |
| pmirGLO-NA-F | TACGAGCTCAGCAAAAGCAGGAGTTTAAAAT |
| pmirGLO-NA-R | CCGCTCGAGTAGTAGAAACAAGGAGTTTTTTGAAC |
| pmirGLO-NP-F | CCGCTCGAGAGCAAAAGCAGGGTAGATAATC |
| pmirGLO-NP-R | TGCTCTAGATAGTAGAAACAAGGGTATTTTTCCT |
| miRNAs | Sequence (5′–3′) |
|---|---|
| ssc-miR-204 mimics Forward | UUCCCUUUGUCAUCCUAUGCCU |
| ssc-miR-204 mimics Reversed | GCAUAGGAUGACAAAGGGAAUU |
| ssc-miR-4331 mimics Forward | UGUGGCUGUGGUGUAGGCCAGC |
| ssc-miR-4331 mimics Reversed | UGGCCUACACCACAGCCACAUU |
| mimics NC Forward | UUCUCCGAACGUGUCACGUTT |
| mimics NC Reversed | ACGUGACACGUUCGGACAATT |
| ssc-miR-204 inhibitor | AGGCAUAGGAUGACAAAGGGAA |
| ssc-miR-4331 inhibitor | GCUGGCCUACACCACAGCCACA |
| inhibitor NC | CAGUACUUUUGUGUAGUACAA |
| Primers | Sequence (5′–3′) |
|---|---|
| HA1-F | GGGTCAAGAAGGGAGAATGA |
| HA1-R | AATGGGAGGCTGGTGTTTAT |
| NS1-F | ACTACTAAGGGCTTTCACTG |
| NS1-R | CATTTCTGCTCTGGAGGT |
| NP-F | CCACAAGAGGGGTCCAGATT |
| NP-R | GGAGATTTCGCTGCACTGAG |
| H5N1-NP-F | CGTTCAGCCCACTTTCTCG |
| H5N1-NP-R | ATCGGGTTCGTTGCCTTTT |
| H9N2-NP-F | AACAGCAGCACAACGAGC |
| H9N2-NP-R | ACAAGCAGGCAAACAGGA |
| GAPDH-F | ACCACAGTCCATGCCATCAC |
| GAPDH-R | TCCACCACCCTGTTGCTGTA |
| ssc-miR-204-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCATA |
| ssc-miR-204-F | TGCGGTTCCCTTTGTCATCCT |
| ssc-miR-204-R | CAGTGCAGGGTCCGAGGT |
| ssc-miR-4331-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTGGCC |
| ssc-miR-4331-F | TGCGGTGTGGCTGTGGTGTAG |
| ssc-miR-4331-R | CAGTGCAGGGTCCGAGGT |
| U6-RT | CGCTTCACGAATTTGCGTGTCAT |
| U6-F | GCTTCGGCAGCACATATACTAAAAT |
| U6-R | CGCTTCACGAATTTGCGTGTCAT |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Wang, R.; Su, H.; Wang, B.; Sizhu, S.; Lei, Z.; Jin, M.; Chen, H.; Cao, J.; Zhou, H. Sus scrofa miR-204 and miR-4331 Negatively Regulate Swine H1N1/2009 Influenza A Virus Replication by Targeting Viral HA and NS, Respectively. Int. J. Mol. Sci. 2017, 18, 749. https://doi.org/10.3390/ijms18040749
Zhang S, Wang R, Su H, Wang B, Sizhu S, Lei Z, Jin M, Chen H, Cao J, Zhou H. Sus scrofa miR-204 and miR-4331 Negatively Regulate Swine H1N1/2009 Influenza A Virus Replication by Targeting Viral HA and NS, Respectively. International Journal of Molecular Sciences. 2017; 18(4):749. https://doi.org/10.3390/ijms18040749
Chicago/Turabian StyleZhang, Shishuo, Ruifang Wang, Huijuan Su, Biaoxiong Wang, Suolang Sizhu, Zhixin Lei, Meilin Jin, Huanchun Chen, Jiyue Cao, and Hongbo Zhou. 2017. "Sus scrofa miR-204 and miR-4331 Negatively Regulate Swine H1N1/2009 Influenza A Virus Replication by Targeting Viral HA and NS, Respectively" International Journal of Molecular Sciences 18, no. 4: 749. https://doi.org/10.3390/ijms18040749






