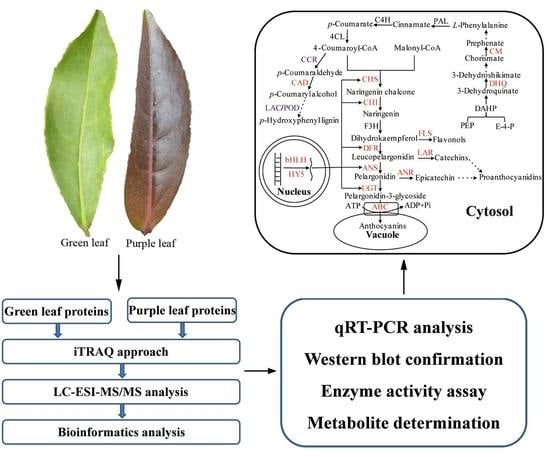
Regulation of Anthocyanin Biosynthesis in Purple Leaves of Zijuan Tea (Camellia sinensis var. kitamura)
Abstract
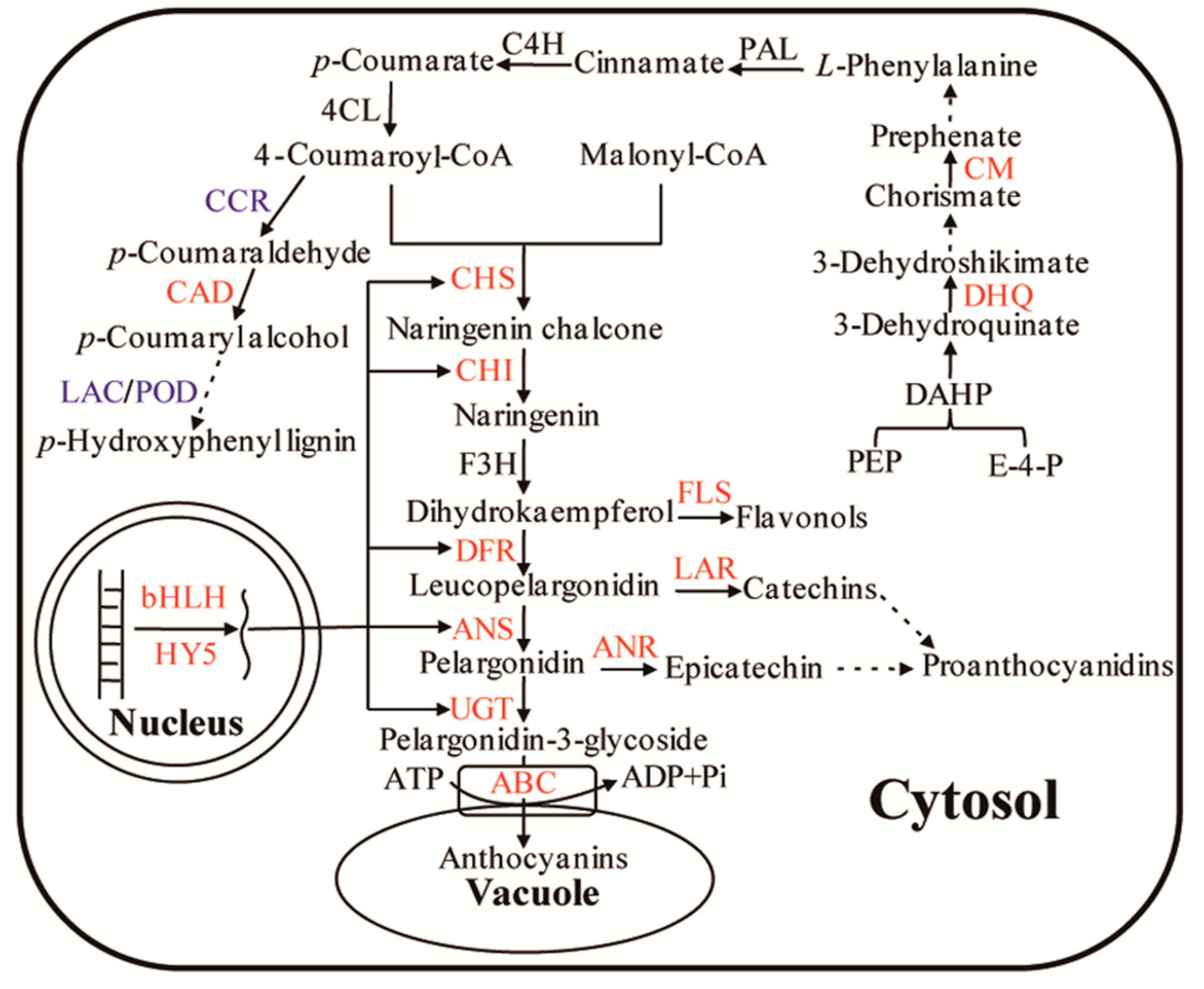
:1. Introduction
2. Results
2.1. Changes in Protein Abundance between Purple and Green Leaves
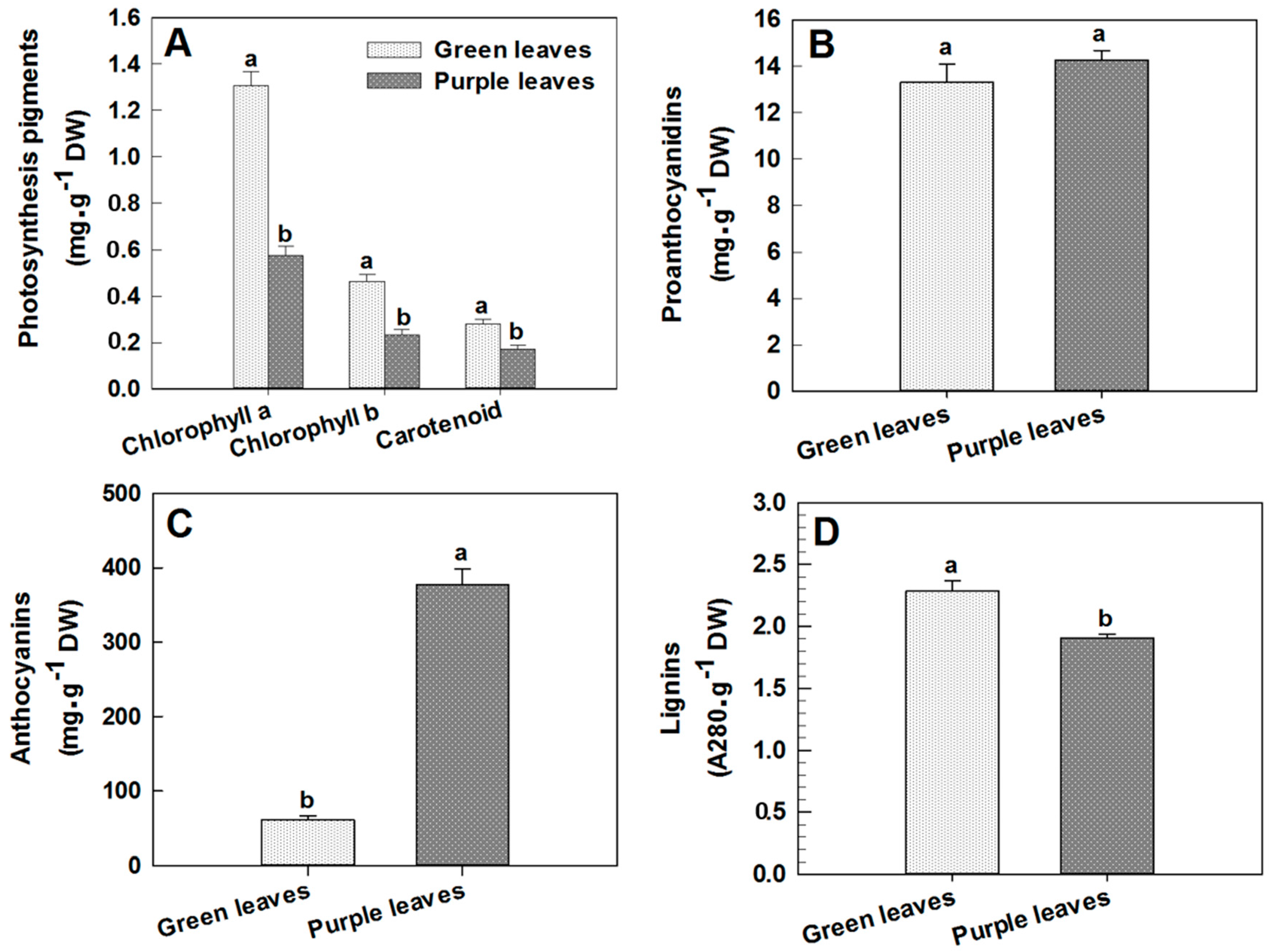
2.2. Changes in Chlorophyll, Proanthocyanidin, Anthocyanin and Lignin Concentrations in Leaves
2.3. Changes in PAL, Chalcone Isomerase (CHI) and UDP-Glycosyl Transferase (UGT) Activities between Purple and Green Leaves
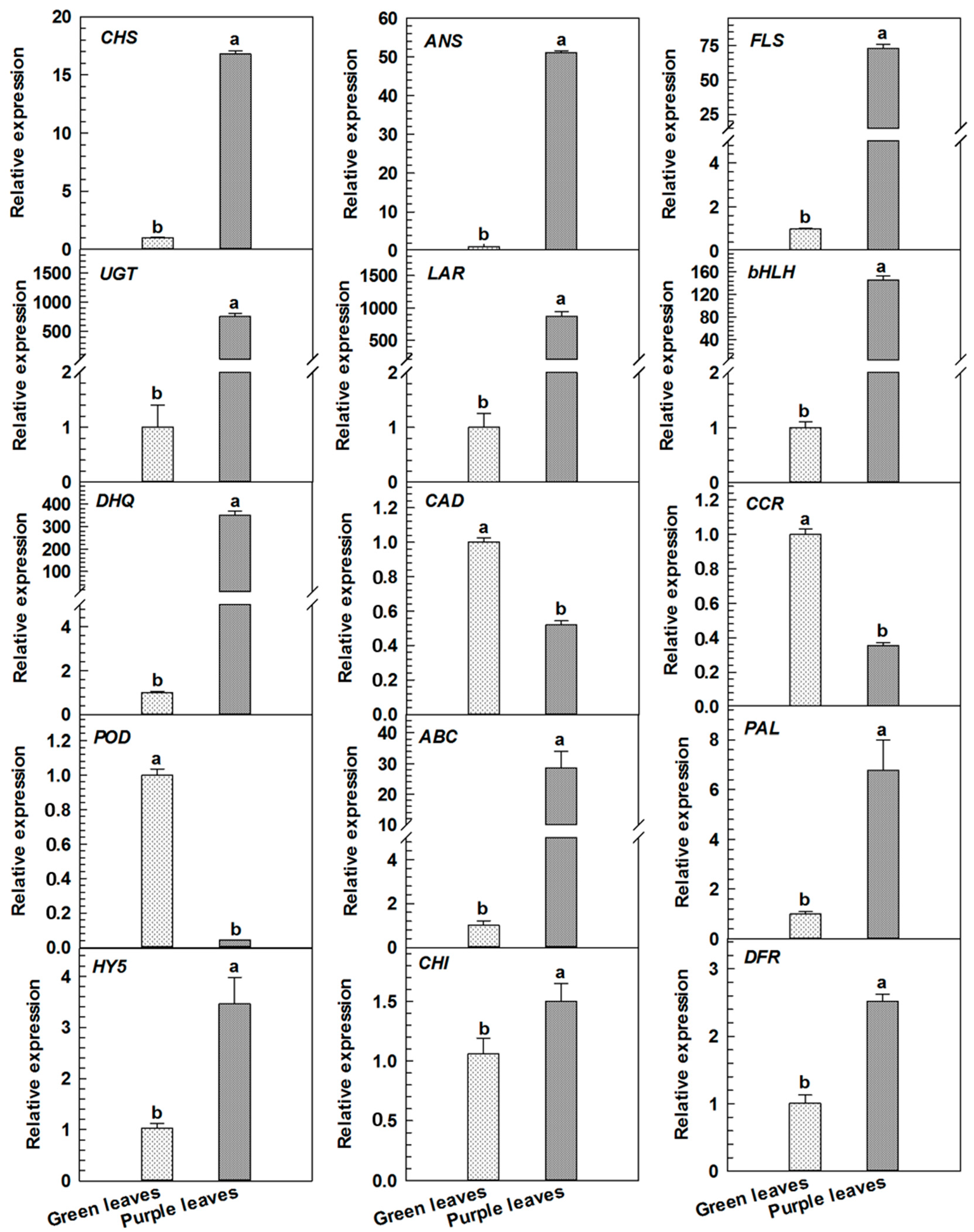
2.4. qRT-PCR Analyses for Differentially Expressed Proteins Related to Anthocyanin Metabolism in Purple Leaves
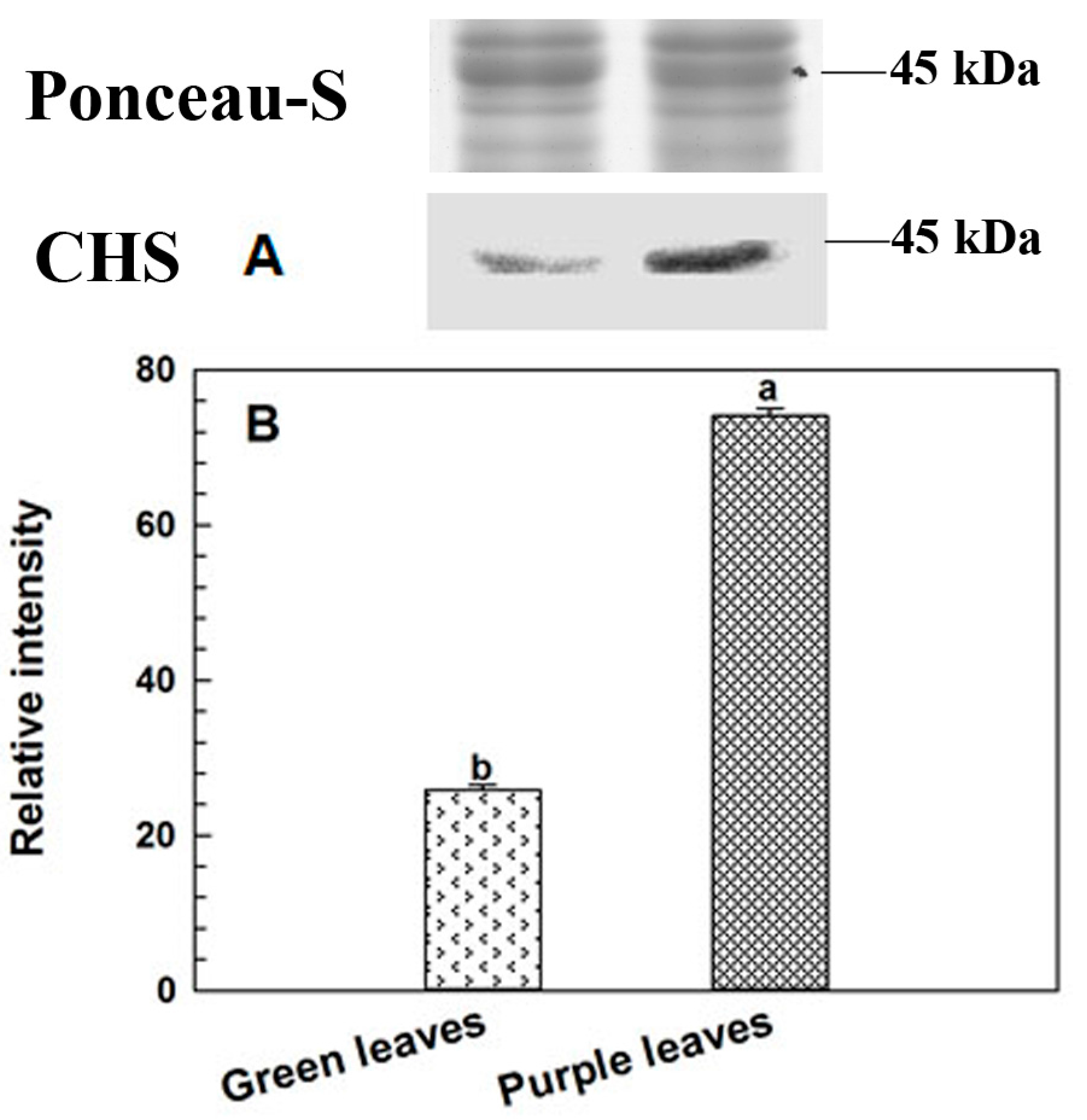
2.5. Immunoblotting Analysis of CHS
3. Discussion
3.1. Regulation of Enzymes and Transporters in Anthocyanin Biosynthesis and Accumulation
3.2. Regulation of Transcription Factors in Anthocyanin Biosynthesis
3.3. Regulation of Substrates in Anthocyanin Biosynthesis
3.4. Regulation of Branched Pathway in Anthocyanin Biosynthesis
4. Materials and Methods
4.1. Plant Materials
4.2. Protein Extraction and iTRAQ Labeling
4.3. Strong Cation Exchange (SCX) Fractionation
4.4. LC-ESI-MS/MS Analysis
4.5. Bioinformatics Analysis
4.6. Chlorophyll, Proanthocyanidin, Anthocyanin, and Lignin Analyses
4.7. PAL, CHI and UGT Assays
4.8. Western Blot Analysis of CHS
4.9. RNA Isolation and qRT-PCR Analysis
4.10. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lin, L.Z.; Sun, J.H.; Chen, P.; Harnly, J.A. LC-PDA-ESI/MSn identification of new anthocyanins in purple Bordeaux radish (Raphanus sativus L. variety). J. Agric. Food Chem. 2011, 59, 6616–6627. [Google Scholar] [CrossRef] [PubMed]
- Neill, S.O.; Gould, K.S.; Kilmartin, P.A.; Mitchell, K.A.; Markham, K.R. Antioxidant activities of red versus green leaves in Elatostema rugosum. Plant Cell Environ. 2002, 25, 539–547. [Google Scholar] [CrossRef]
- Kerio, L.C.; Wachira, F.N.; Wanyoko, J.K.; Rotich, M.K. Total polyphenols, catechin profiles and antioxidant activity of tea products from purple leaf coloured tea cultivars. Food Chem. 2013, 136, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Xu, J.; Kim, J.; Chen, T.Y.; Su, X.; Standard, J.; Carey, E.; Griffin, J.; Herndon, B.; Katz, B.; et al. Role of anthocyanin-enriched purple-fleshed sweet potato p40 in colorectal cancer prevention. Mol. Nutr. Food Res. 2013, 57, 1908–1917. [Google Scholar] [CrossRef] [PubMed]
- Forester, S.C.; Choy, Y.Y.; Waterhouse, A.L.; Oteiza, P.I. The anthocyanin metabolites gallic acid, 3-O-methylgallic acid, and 2,4,6-trihydroxybenzaldehyde decrease human colon cancer cell viability by regulating pro-oncogenic signals. Mol. Carcinog. 2014, 53, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhao, Y.; Tao, X.Y.; Zhang, M.; Sun, A.D. Protective effect of blueberry anthocyanins in a CCl4-induced liver cell model. Food Sci. Technol. 2015, 60, 1105–1112. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; Leppen, H.T.C.; Mol, J.N.M.; Koes, R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 1993, 5, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Wrolstad, R.E. Anthocyanin pigments-Bioactivity and coloring properties. J. Food Sci. 2004, 69, C419–C425. [Google Scholar] [CrossRef]
- Jiang, L.; Shen, X.; Shoji, T.; Kanda, T.; Zhou, J.; Zhao, L. Characterization and activity of anthocyanins in Zijuan tea (Camellia sinensis var. kitamura). J. Agric. Food Chem. 2013, 61, 3306–3310. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.P.; Dai, W.D.; Tan, J.F.; Guo, L.; Zhu, Y.; Lin, Z. Identification of the anthocyanins from the purple leaf coloured tea cultivar Zijuan (Camellia sinensis var. assamica) and characterization of their antioxidant activities. J. Funct. Foods 2015, 17, 449–458. [Google Scholar]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef] [PubMed]
- Kroon, J.; Souer, E.; Graaff, A.; Xue, Y.; Mol, J.; Koes, R. Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: Characterization of insertion sequences in two mutant alleles. Plant J. 1994, 5, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar transparent testa glabra1 gene in Arabidopsis thaliana. Plant Cell 2004, 16, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Kerio, L.C.; Wachira, F.N.; Wanyoko, J.K.; Rotich, M.K. Characterization of anthocyanins in Kenyan teas: Extraction and identification. Food Chem. 2012, 131, 31–38. [Google Scholar] [CrossRef]
- Saito, T.; Honma, D.; Tagashira, M.; Kanda, T.; Nesumi, A.; Maeda-Yamamoto, M. Anthocyanins from new red leaf tea ‘Sunrouge’. J. Agric. Food Chem. 2011, 59, 4779–4782. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lv, X.; Wang, L.; Qiu, Z.; Song, X.; Lin, J.; Chen, W. Transcriptome analysis reveals the accumulation mechanism of anthocyanins in ‘Zijuan’ tea (Camellia sinensis, var. asssamica, (Masters) kitamura) leaves. Plant Growth Regul. 2017, 81, 51–61. [Google Scholar] [CrossRef]
- Wang, Z.W.; Jiang, C.; Wen, Q.; Wang, N.; Tao, Y.Y.; Xu, L.A. Deep sequencing of the Camellia chekiangoleosa transcriptome revealed candidate genes for anthocyanin biosynthesis. Gene 2014, 538, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, J.B.; Hahn, B.S.; Kim, K.H.; Ha, S.H.; Kim, J.B.; Kim, Y.H. EST analysis of genes involved in secondary metabolism in Camellia sinensis (tea), using suppression subtractive hybridization. Plant Sci. 2004, 166, 953–961. [Google Scholar] [CrossRef]
- Yang, D.; Liu, Y.; Sun, M.; Zhao, L.; Wang, Y.; Chen, X.; Wei, C.; Gao, L.; Xia, T. Differential gene expression in tea (Camellia sinensis L.) calli with different morphologies and catechin contents. J. Plant Physiol. 2012, 169, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Van der Krol, A.R.; Lenting, P.E.; Veenstra, J.; Van der Meer, I.M.; Koes, R.E.; Gerats, A.G.M.; Mol, J.N.M.; Stuitje, A.R. An anti-sense chalcone synthase gene in transgenetic plants inhibits flower pigmentation. Nature 1988, 333, 860–869. [Google Scholar] [CrossRef]
- Fukusaki, E.I.; Kawasaki, K.; Kajiyama, S.I.; An, C.I.; Suzuki, K.; Tanaka, Y.; Kobayashi, A. Flower color modulations of Torenia hybrida by down regulation of chalcone synthase genes with RNA interference. J. Biotechnol. 2004, 111, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Halbwirth, H.; Puhl, I.; Haas, U.; Jezik, K.; Treutter, D.; Stich, K. Two-phase flavonoid formation in developing strawberry (Fragaria × ananassa) fruit. J. Agric. Food Chem. 2006, 54, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.M.; Reddy, V.S.; Scheffler, B.E.; Wienand, U.; Reddy, A.R. Novel transgenic rice overexpressing anthocyanidin synthase accumulates a mixture of flavonoids leading to an increased antioxidant potential. Metab. Eng. 2007, 9, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Annilo, T. Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu. Rev. Genom. Hum. Genet. 2005, 6, 123–142. [Google Scholar] [CrossRef] [PubMed]
- Braidot, E.; Zancani, M.; Petrussa, E.; Peresson, C.; Bertolini, A.; Patui, S.; Macri, F.; Vianello, A. Transport and accumulation of flavonoids in grapevine (Vitis vinifera L.). Plant Signal. Behav. 2008, 3, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, M.; Meng, X.; Wan, H.; Zhang, J.; Tian, J.; Hao, S.; Jin, K.; Yao, Y. Photoperiod and shading regulate coloration and anthocyanin accumulation in the leaves of malus crabapples. Plant Cell Tissue Organ Cult. 2015, 121, 619–632. [Google Scholar] [CrossRef]
- Liu, X.F.; Yin, X.R.; Allan, A.C.; Lin-Wang, K.; Shi, Y.N.; Huang, Y.J.; Ferguson, I.B.; Chen, K.S. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tissue Organ 2013, 115, 285–298. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Zhu, Z.; Cao, P.; Hao, C.; Chen, C.; Xin, Z.; Mao, Y.; Lei, J.; Jiang, Y.; Meng, W.; et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016, 6, 32534. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; He, K.; Stolc, V.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, H.; Wang, X.; Wang, X.; Yang, X.; Li, L.; Deng, X.W. Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-translational regulation. Plant J. 2011, 65, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.H.; Choi, M.G.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547. [Google Scholar] [CrossRef] [PubMed]
- Knaggs, A.R. The biosynthesis of shikimate metabolites. Nat. Prod. Rep. 2003, 20, 119–136. [Google Scholar] [CrossRef] [PubMed]
- Chabannes, M.; Barakate, A.; Lapierre, C.; Marita, J.M.; Ralph, J.; Pean, M.; Danoun, S.; Halpin, C.; Grima-Pettenati, J.; Boudet, A.M. Strong decrease in lignin content without significant alteration of plant development is induced by simultaneous down-regulation of cinnamoyl CoA reductase (CCR) and cinnamyl alcohol dehydrogenase (CAD) in tobacco plants. Plant J. 2001, 28, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Leplé, J.C.; Dauwe, R.; Morreel, K.; Storme, V.; Lapierre, C.; Pollet, B.; Naurmann, A.; Kang, K.Y.; Kim, H.; Ruel, K.; et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on dell wall polymer metabolism and structure. Plant Cell 2007, 19, 3669–3691. [Google Scholar] [CrossRef] [PubMed]
- Lapierre, C.; Pollet, P.; Petit-Conil, M.; Toval, G.; Romero, J.; Pilate, G.; Leplé, J.C.; Boerjan, W.; Ferret, V.; de Nadai, V.; et al. Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or daffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial kraft pulping. Plant Physiol. 1999, 119, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Pilate, G.; Guiney, E.; Holt, K.; Petit-Conil, M.; Lapierre, C.; Leplé, J.C.; Pollet, B.; Mila, I.; Webster, E.A.; Marstorp, H.G.; et al. Field and pulping performances of transgenic trees with altered lignification. Nat. Biotechnol. 2002, 20, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y.; Demura, T.; Yamawaki, K.; Inoue, Y.; Sato, S.; Sugiyama, M.; Fukuda, H. Isolation and characterization of a novel peroxidase gene ZPO-C whose expression and function are closely associated with lignification during tracheary element differentiation. Plant Cell Physiol. 2006, 47, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Davis, E.; Gardner, D.; Cai, X.; Wu, Y. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 2006, 224, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Xia, L.; Li, Y.; Liang, M. A new tea tree cultivar “Zijuan”. Acta Hortic. 2008, 35, 934. [Google Scholar]
- Liu, H.; Liu, Y.Z.; Zheng, S.Q.; Jiang, J.M.; Wang, P.; Chen, W. Comparative proteomic analysis of longan (Dimocarpus longan Lour.) seed abortion. Planta 2010, 231, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophyll and carotenoids: Pigments of photosynthetic biomembranes. Meth. Enzymol. 1987, 148, 331–382. [Google Scholar]
- Skerget, M.; Kotnik, P.; Hadolin, M.; Hraš, A.R.; Simonič, M.; Knez, Ž. Phenols, proanthocyanidins, flavones and flavonols in some plant materials and their antioxidant activities. Food Chem. 2005, 89, 191–198. [Google Scholar] [CrossRef]
- Zheng, X.; Tian, S. Effect of oxalic acid on control of postharvest browning of litchi fruit. Food Chem. 2006, 96, 519–523. [Google Scholar] [CrossRef]
- Syros, T.; Yupsanis, T.; Zafiriadis, H.; Economou, A. Activity and isoforms of peroxidases, lignin and anatomy, during adventitious rooting in cuttings of Ebenus cretica L. J. Plant Physiol. 2004, 161, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Zhang, S.; Xiao, C.; Zhang, Q.; Qin, G.; Wu, J. Relationship between anthocyanin biosynthesis and related enzyme activities in Pyrus communis L. cv. “Early Red Comic”’ and its green bud mutant. Acta Bot. Boreal. Occident. Sin. 2011, 31, 1428–1433. [Google Scholar]
- Wang, L.; Liang, W.; Xing, J.H.; Tan, F.; Chen, Y.; Huang, L.; Cheng, C.-L.; Chen, W. Dynamics of chloroplast proteome in salt-stressed mangrove Kandelia candel (L.) Druce. J. Proteom. Res. 2013, 12, 5124–5136. [Google Scholar] [CrossRef] [PubMed]
- Jordon-Thaden, I.E.; Chanderbali, A.S.; Gitzendanner, M.A.; Soltis, D.E. Modified CTAB and TRIzol protocols improve RNA extraction from chemically complex Embryophyta. Appl. Plant Sci. 2015, 3, a1400105. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| No. | Protein Name a | Sequence Name b | Accession No. c | E-Value d | Similarity | G e | P f | Fold |
|---|---|---|---|---|---|---|---|---|
| Anthocyanin metabolism | ||||||||
| 1 | chalcone synthase | c38979.graph_c0 | gi|320117902 | 0 | 94.45% | 1 | 3 | 3 |
| 2 | chalcone isomerase | c22272.graph_c0 | gi|297736495 | 5.23 × 10−121 | 91.30% | 1 | 2.7 | 2.7 |
| 3 | chalcone isomerase | c38282.graph_c0 | gi|325551315 | 9.70 × 10−149 | 90.75% | 1 | 2.3 | 2.3 |
| 4 | dihydroflavonol 4-reductase | c30221.graph_c0 | gi|6009511 | 0 | 93.25% | 1 | 1.5 | 1.5 |
| 5 | anthocyanidin synthase | c13490.graph_c0 | gi|378749118 | 0 | 92.50% | 1 | 1.8 | 1.8 |
| 6 | UDP-glucosyl transferase 88A1 | c41584.graph_c0 | gi|508711676 | 5.32 × 10−153 | 71.00% | 1 | 2.6 | 2.6 |
| 7 | abc transporter b family member 8 | c30601.graph_c0 | gi|359484339 | 0 | 90.00% | 1 | 1.8 | 1.8 |
| 8 | flavonol synthase/flavanone 3-hydroxylase-like | c30094.graph_c0 | gi|76786311 | 0 | 88.30% | 1 | 2.4 | 2.4 |
| 9 | leucoanthocyanidin reductase | c29035.graph_c0 | gi|326380568 | 0 | 86.55% | 1 | 3.4 | 3.4 |
| 10 | anthocyanidin reductase | c36734.graph_c0 | gi|294847480 | 0 | 93.10% | 1 | 2.4 | 2.4 |
| Transcription factor | ||||||||
| 11 | transcription factor bhlh135 | c13939.graph_c0 | gi|508711524 | 3.92 × 10−47 | 92.50% | 1 | 1.7 | 1.7 |
| 12 | transcription factor bhlh66-like | c38737.graph_c0 | gi|508782323 | 1.30 × 10−78 | 67.20% | 1 | 2.2 | 2.2 |
| 13 | transcription factor hy5 | c31532.graph_c0 | gi|470110394 | 6.94 × 10−57 | 87.45% | 1 | 2.5 | 2.5 |
| Shikimic acid pathway | ||||||||
| 14 | bifunctional 3-dehydroquinate dehydratase shikimate chloroplastic-like | c27480.graph_c0 | gi|259479224 | 0 | 87.55% | 1 | 1.5 | 1.5 |
| 15 | bifunctional 3-dehydroquinate dehydratase shikimate chloroplastic-like | c29706.graph_c0 | gi|225451146 | 0 | 84.15% | 1 | 2.2 | 2.2 |
| 16 | chorismate mutase chloroplastic | c24623.graph_c0 | gi|460372757 | 1.24 × 10−44 | 70.80% | 1 | 1.8 | 1.8 |
| Lignin synthesis | ||||||||
| 17 | Predicted: Cinnamoyl-CoA reductase 1 | c41372.graph_c0 | gi|225452438 | 0 | 91.05% | 1 | 0.6 | 0.6 |
| 18 | cinnamyl alcohol dehydrogenase | c34762.graph_c0 | gi|332384181 | 0 | 91.80% | 1 | 1.6 | 1.6 |
| 19 | Laccase-6 (Precursor) | c35053.graph_c0 | gi|297737720 | 0 | 85.90% | 1 | 0.4 | 0.4 |
| 20 | peroxidase 15-like | c41476.graph_c1 | gi|462402484 | 4.81 × 10−147 | 76.85% | 1 | 0.4 | 0.4 |
| Genes | Primer Sequence(5′→3′) |
|---|---|
| Actin | GGCAGATAGATGCTTATGTAGGTG |
| TGTTTGCTTTAGGGTTGAGTGG | |
| CHS | AGTGGAGGAAGTGAGGAGGG |
| CGCTGTTAGTAATGCGGAAGT | |
| CHI | TCCAAGCCCTTCTTCCTCG |
| GACCCGTGAATGGCAAAATC | |
| DFR | AGAGCAGGGAGGCTTGTATG |
| GAGTATTTGGACCGATGTGG | |
| FLS | ATACAGGGGAGTGACAGAGGAAT |
| CATTGGGGACAAGTAAAGTGAGA | |
| ANS | ACGAGGGCAAATGGGTCA |
| TCCTTGGGTGGTTCGCAGA | |
| UGT | AATCTGTGGTGTCCGTTTGCTTC |
| TCTTCGCTGTCTTCTTTGTCTACTT | |
| LAR | TGCGGCGATTGATAGAAG |
| GCAGGATGGTCGGAAATG | |
| bHLH | CCCGAGATTCGCAATAGGC |
| TGTCGCTGAGATCATCCACTTC | |
| POD | CGGGCTGGATGCTTGACTGT |
| CCCTGCTTGTTCTGGAGGTTAG | |
| CCR | AGCAATGGTTGTCGGTCCTC |
| ACCTGGTAAGTCGGCGTAAAT | |
| CAD | CCTCGGTCCAGACGATTACG |
| CAACAAAGAAAGGTATGGCTCAAGT | |
| DHQ | GAAGCCTGAAAAGGTCAAAATC |
| TTGGCACAAAGTATGCGAGA | |
| PAL | ACACTTTATGTGCCCAAGACCC |
| GCTTCCGATACTCCGCTACCA | |
| ABC | GGTTATGCTGAGCCTGGTAGT |
| CCGTTGAGGAGAATAGTGCC | |
| HY5 | AGGGTCGGTGTCTTTCAG |
| GTATGCCTTCTTCCTTTCC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Pan, D.; Liang, M.; Abubakar, Y.S.; Li, J.; Lin, J.; Chen, S.; Chen, W. Regulation of Anthocyanin Biosynthesis in Purple Leaves of Zijuan Tea (Camellia sinensis var. kitamura). Int. J. Mol. Sci. 2017, 18, 833. https://doi.org/10.3390/ijms18040833
Wang L, Pan D, Liang M, Abubakar YS, Li J, Lin J, Chen S, Chen W. Regulation of Anthocyanin Biosynthesis in Purple Leaves of Zijuan Tea (Camellia sinensis var. kitamura). International Journal of Molecular Sciences. 2017; 18(4):833. https://doi.org/10.3390/ijms18040833
Chicago/Turabian StyleWang, Lingxia, Dezhuo Pan, Meng Liang, Yakubu Saddeeq Abubakar, Jian Li, Jinke Lin, Shipin Chen, and Wei Chen. 2017. "Regulation of Anthocyanin Biosynthesis in Purple Leaves of Zijuan Tea (Camellia sinensis var. kitamura)" International Journal of Molecular Sciences 18, no. 4: 833. https://doi.org/10.3390/ijms18040833