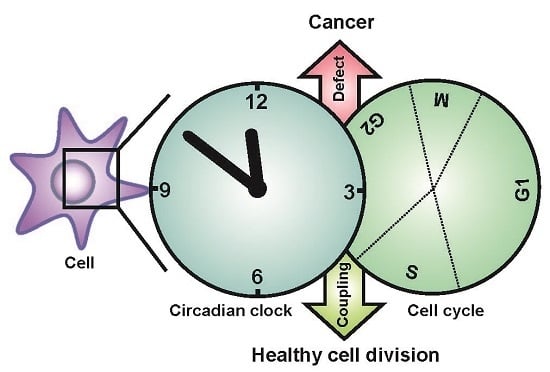
Circadian Clock, Cell Division, and Cancer: From Molecules to Organism
Abstract
:1. Introduction
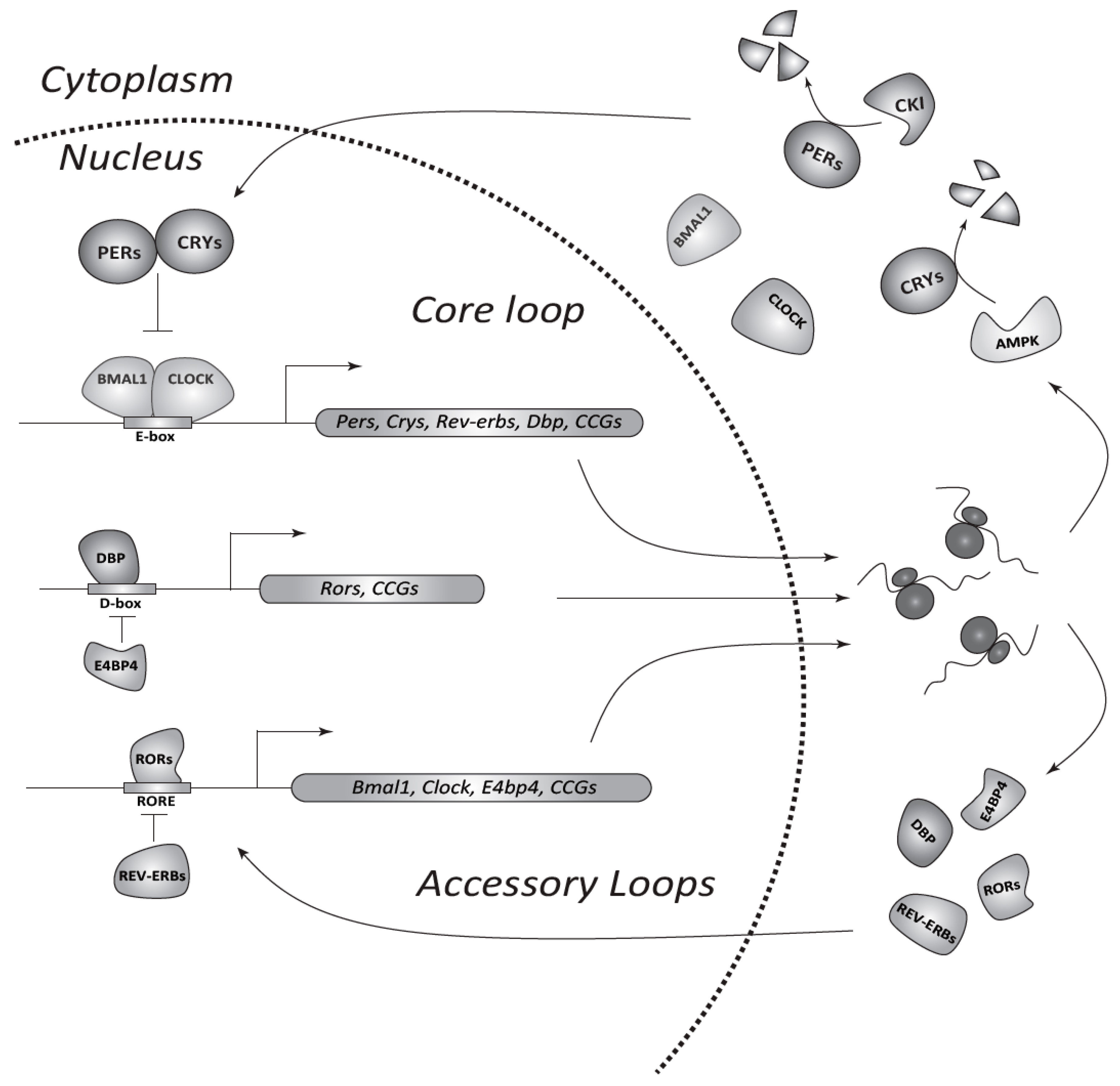
2. The Circadian System in Mammals
3. The Cell Cycle in Mammals
4. Molecular Links between the Circadian Clock and the Cell Cycle
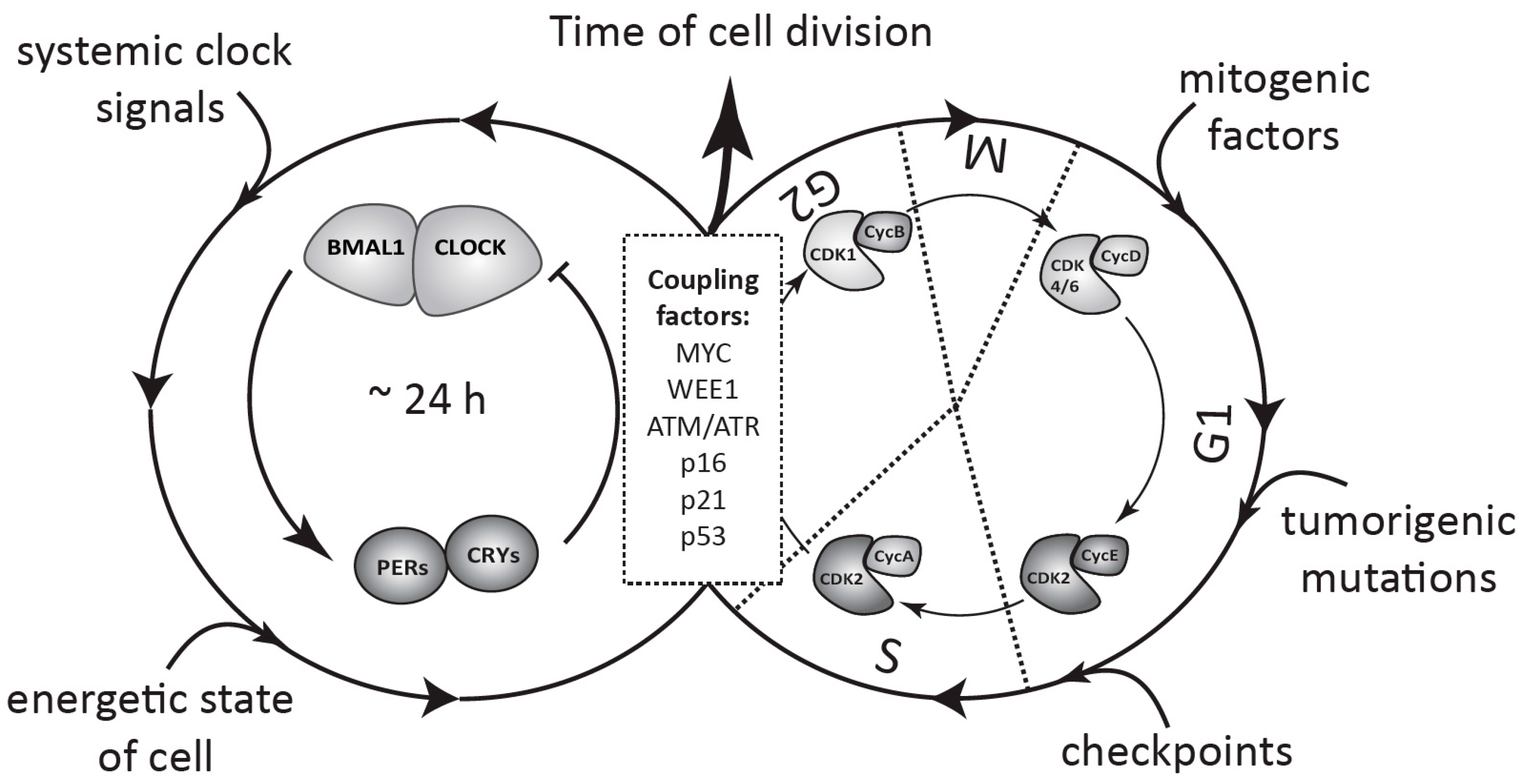
5. Coupling between the Circadian Clock and the Cell Cycle
6. Physiological Significance of the Clock-Cell Cycle Coupling
7. The Circadian Clock as a Tumor Suppressor
8. Does the Circadian Clock Support Tumorigenesis?
9. Cancer Affects Circadian Rhythms in Cells and in the Body
10. Conclusions
Acknowledgments
Conflicts of Interest
References
- Bass, J. Circadian topology of metabolism. Nature 2012, 491, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Woelfle, M.A.; Ouyang, Y.; Phanvijhitsiri, K.; Johnson, C.H. The adaptive value of circadian clocks: An experimental assessment in cyanobacteria. Curr. Biol. 2004, 14, 1481–1486. [Google Scholar] [CrossRef] [PubMed]
- Rosbash, M. The implications of multiple circadian clock origins. PLoS Biol. 2009, 7, e62. [Google Scholar] [CrossRef] [PubMed]
- Pittendrigh, C.S. Temporal organization: Reflections of a darwinian clock-watcher. Annu. Rev. Physiol. 1993, 55, 16–54. [Google Scholar] [CrossRef] [PubMed]
- Khapre, R.V.; Samsa, W.E.; Kondratov, R.V. Circadian regulation of cell cycle: Molecular connections between aging and the circadian clock. Ann. Med. 2010, 42, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Lowrey, P.L.; Takahashi, J.S. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genom. Hum. Genet. 2004, 5, 407–441. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H. Circadian clocks and cell division: What’s the pacemaker? Cell Cycle 2010, 9, 3864–3873. [Google Scholar] [CrossRef] [PubMed]
- Geyfman, M.; Kumar, V.; Liu, Q.; Ruiz, R.; Gordon, W.; Espitia, F.; Cam, E.; Millar, S.E.; Smyth, P.; Ihler, A.; et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc. Natl. Acad. Sci. USA 2012, 109, 11758–11763. [Google Scholar] [CrossRef] [PubMed]
- Gaddameedhi, S.; Selby, C.P.; Kaufmann, W.K.; Smart, R.C.; Sancar, A. Control of skin cancer by the circadian rhythm. Proc. Natl. Acad. Sci. USA 2011, 108, 18790–18795. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, S.S.; Johnson, C.H. Daily and circadian variation in survival from ultraviolet radiation in chlamydomonas reinhardtii. Photochem. Photobiol. 2000, 71, 758–765. [Google Scholar] [CrossRef]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.J.; Liu, D. Cryptochromes: Blue light receptors for plants and animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Gehring, W.; Rosbash, M. The coevolution of blue-light photoreception and circadian rhythms. J. Mol. Evol. 2003, 57, S286–S289. [Google Scholar] [CrossRef] [PubMed]
- Oklejewicz, M.; Destici, E.; Tamanini, F.; Hut, R.A.; Janssens, R.; van der Horst, G.T. Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol. 2008, 18, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, B.M. Resetting the biological clock in gonyaulax with ultraviolet light. Plant Physiol. 1963, 38, 704–708. [Google Scholar] [CrossRef] [PubMed]
- Tilbrook, K.; Arongaus, A.B.; Binkert, M.; Heijde, M.; Yin, R.; Ulm, R. The UVR8 UV-B photoreceptor: Perception, signaling and response. Arabidopsis Book 2013, 11, e0164. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.B.; Tossi, V.; Lamattina, L.; Cassia, R. A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front. Plant Sci. 2016, 7, 1698. [Google Scholar] [CrossRef] [PubMed]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Feher, B.; Kozma-Bognar, L.; Kevei, E.; Hajdu, A.; Binkert, M.; Davis, S.J.; Schafer, E.; Ulm, R.; Nagy, F. Functional interaction of the circadian clock and UV resistance locus 8-controlled UV-B signaling pathways in arabidopsis thaliana. Plant J. Cell Mol. Biol. 2011, 67, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Tu, B.P.; Kudlicki, A.; Rowicka, M.; McKnight, S.L. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science 2005, 310, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Causton, H.C.; Feeney, K.A.; Ziegler, C.A.; O’Neill, J.S. Metabolic cycles in yeast share features conserved among circadian rhythms. Curr. Biol. 2015, 25, 1056–1062. [Google Scholar] [CrossRef] [PubMed]
- Mellor, J. The molecular basis of metabolic cycles and their relationship to circadian rhythms. Nat. Struct. Mol. Biol. 2016, 23, 1035–1044. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Odstrcil, E.A.; Tu, B.P.; McKnight, S.L. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science 2007, 316, 1916–1919. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; McKnight, S.L. A conserved DNA damage response pathway responsible for coupling the cell division cycle to the circadian and metabolic cycles. Cell Cycle 2007, 6, 2906–2912. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.M.; Loros, J.J.; Dunlap, J.C. Circadian oscillators: Around the transcription-translation feedback loop and on to output. Trends Biochem. Sci. 2016, 41, 834–846. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.E.; Kay, S.A. The plant circadian clock: From a simple timekeeper to a complex developmental manager. Cold Spring Harb. Perspect. Biol. 2016, 8, a027748. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, J.S. Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 2016, 18, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Lahens, N.F.; Ballance, H.I.; Hughes, M.E.; Hogenesch, J.B. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc. Natl. Acad. Sci. USA 2014, 111, 16219–16224. [Google Scholar] [CrossRef] [PubMed]
- Robles, M.S.; Humphrey, S.J.; Mann, M. Phosphorylation is a central mechanism for circadian control of metabolism and physiology. Cell Metab. 2017, 25, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.G.; Dubus, P.; Mettus, R.V.; Galbreath, E.J.; Boden, G.; Reddy, E.P.; Barbacid, M. Loss of CDK4 expression causes insulin-deficient diabetes and CDK4 activation results in beta-islet cell hyperplasia. Nat. Genet. 1999, 22, 44–52. [Google Scholar] [PubMed]
- Malumbres, M.; Sotillo, R.; Santamaria, D.; Galan, J.; Cerezo, A.; Ortega, S.; Dubus, P.; Barbacid, M. Mammalian cells cycle without the D-type cyclin-dependent kinases CDK4 and CDK6. Cell 2004, 118, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Berthet, C.; Aleem, E.; Coppola, V.; Tessarollo, L.; Kaldis, P. Cdk2 knockout mice are viable. Curr. Biol. 2003, 13, 1775–1785. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Prieto, I.; Odajima, J.; Martin, A.; Dubus, P.; Sotillo, R.; Barbero, J.L.; Malumbres, M.; Barbacid, M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 2003, 35, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, D.; Barriere, C.; Cerqueira, A.; Hunt, S.; Tardy, C.; Newton, K.; Caceres, J.F.; Dubus, P.; Malumbres, M.; Barbacid, M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007, 448, 811–815. [Google Scholar] [CrossRef] [PubMed]
- Curtin, N.J. DNA repair dysregulation from cancer driver to therapeutic target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, T.; Yamaguchi, S.; Mitsui, S.; Emi, A.; Shimoda, F.; Okamura, H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 2003, 302, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Zhao, S.; Jiang, X.; Zhang, E.; Hu, G.; Hu, B.; Zheng, P.; Xiao, J.; Lu, Z.; Lu, Y.; et al. The circadian clock gene bmal1 acts as a potential anti-oncogene in pancreatic cancer by activating the p53 tumor suppressor pathway. Cancer Lett. 2016, 371, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Kowalska, E.; Ripperger, J.A.; Hoegger, D.C.; Bruegger, P.; Buch, T.; Birchler, T.; Mueller, A.; Albrecht, U.; Contaldo, C.; Brown, S.A. Nono couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. USA 2013, 110, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Grechez-Cassiau, A.; Rayet, B.; Guillaumond, F.; Teboul, M.; Delaunay, F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 2008, 283, 4535–4542. [Google Scholar] [CrossRef] [PubMed]
- Papp, S.J.; Huber, A.L.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Moresco, J.J.; Yates, J.R.; Lamia, K.A. DNA damage shifts circadian clock time via Hausp-dependent Cry1 stabilization. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Miyagishima, S.Y.; Fujiwara, T.; Sumiya, N.; Hirooka, S.; Nakano, A.; Kabeya, Y.; Nakamura, M. Translation-independent circadian control of the cell cycle in a unicellular photosynthetic eukaryote. Nat. Commun. 2014, 5, 3807. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.L.; Papp, S.J.; Chan, A.B.; Henriksson, E.; Jordan, S.D.; Kriebs, A.; Nguyen, M.; Wallace, M.; Li, Z.; Metallo, C.M.; et al. CRY2 and FBXL3 cooperatively degrade c-MYC. Mol. Cell 2016, 64, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Vila-Caballer, M.; Liu, J.; Schiffhauer, S.; Finkielstein, C.V. Association of the circadian factor period 2 to p53 influences p53’s function in DNA-damage signaling. Mol. Biol. Cell 2015, 26, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Vila-Caballer, M.; Santos, C.S.; Liu, J.; Yang, J.; Finkielstein, C.V. The circadian factor period 2 modulates p53 stability and transcriptional activity in unstressed cells. Mol. Biol. Cell 2014, 25, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Kim, J.K.; Liu, J.; Vila-Caballer, M.; Stauffer, P.E.; Tyson, J.J.; Finkielstein, C.V. Model-driven experimental approach reveals the complex regulatory distribution of p53 by the circadian factor period 2. Proc. Natl. Acad. Sci. USA 2016, 113, 13516–13521. [Google Scholar] [CrossRef] [PubMed]
- Gery, S.; Komatsu, N.; Baldjyan, L.; Yu, A.; Koo, D.; Koeffler, H.P. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol. Cell 2006, 22, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Gotter, A.L.; Manganaro, T.; Weaver, D.R.; Kolakowski, L.F., Jr.; Possidente, B.; Sriram, S.; MacLaughlin, D.T.; Reppert, S.M. A time-less function for mouse timeless. Nat. Neurosci. 2000, 3, 755–756. [Google Scholar] [PubMed]
- Gotter, A.L. A timeless debate: Resolving tim’s noncircadian roles with possible clock function. Neuroreport 2006, 17, 1229–1233. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.W.; Tischkau, S.A.; Barnes, J.A.; Mitchell, J.W.; Burgoon, P.W.; Hickok, J.R.; Gillette, M.U. Requirement of mammalian timeless for circadian rhythmicity. Science 2003, 302, 439–442. [Google Scholar] [CrossRef] [PubMed]
- Unsal-Kacmaz, K.; Mullen, T.E.; Kaufmann, W.K.; Sancar, A. Coupling of human circadian and cell cycles by the timeless protein. Mol. Cell. Biol. 2005, 25, 3109–3116. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wood, P.A.; Hrushesky, W.J. Mammalian TIMELESS is required for ATM-dependent CHK2 activation and G2/M checkpoint control. J. Biol. Chem. 2010, 285, 3030–3034. [Google Scholar] [CrossRef] [PubMed]
- Feillet, C.; van der Horst, G.T.; Levi, F.; Rand, D.A.; Delaunay, F. Coupling between the circadian clock and cell cycle oscillators: Implication for healthy cells and malignant growth. Front. Neurol. 2015, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Mori, T.; Binder, B.; Johnson, C.H. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc. Natl. Acad. Sci. USA 1996, 93, 10183–10188. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Yang, Q.; Wang, Q.; Kim, Y.I.; Wood, T.L.; Osteryoung, K.W.; van Oudenaarden, A.; Golden, S.S. Elevated atpase activity of kaic applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 2010, 140, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Buchi, K.N.; Moore, J.G.; Hrushesky, W.J.; Sothern, R.B.; Rubin, N.H. Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology 1991, 101, 410–415. [Google Scholar] [CrossRef]
- Scheving, L.E.; Burns, E.R.; Pauly, J.E.; Tsai, T.H. Circadian variation in cell division of the mouse alimentary tract, bone marrow and corneal epithelium. Anat. Rec. 1978, 191, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Smaaland, R.; Laerum, O.D.; Lote, K.; Sletvold, O.; Sothern, R.B.; Bjerknes, R. DNA synthesis in human bone marrow is circadian stage dependent. Blood 1991, 77, 2603–2611. [Google Scholar] [PubMed]
- Bjarnason, G.A.; Jordan, R. Rhythms in human gastrointestinal mucosa and skin. Chronobiol. Int. 2002, 19, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Nagoshi, E.; Saini, C.; Bauer, C.; Laroche, T.; Naef, F.; Schibler, U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell 2004, 119, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Bieler, J.; Cannavo, R.; Gustafson, K.; Gobet, C.; Gatfield, D.; Naef, F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol. Syst. Biol. 2014, 10, 739. [Google Scholar] [CrossRef] [PubMed]
- Feillet, C.; Krusche, P.; Tamanini, F.; Janssens, R.C.; Downey, M.J.; Martin, P.; Teboul, M.; Saito, S.; Levi, F.A.; Bretschneider, T.; et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc. Natl. Acad. Sci. USA 2014, 111, 9828–9833. [Google Scholar] [CrossRef] [PubMed]
- Matsu-Ura, T.; Dovzhenok, A.; Aihara, E.; Rood, J.; Le, H.; Ren, Y.; Rosselot, A.E.; Zhang, T.; Lee, C.; Obrietan, K.; et al. Intercellular coupling of the cell cycle and circadian clock in adult stem cell culture. Mol. Cell 2016, 64, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Dawson, K.B.; Madoc-Jones, H.; Field, E.O. Variations in the generation times of a strain of rat sarcoma cells in culture. Exp. Cell Res. 1965, 38, 75–84. [Google Scholar] [CrossRef]
- Sandler, O.; Mizrahi, S.P.; Weiss, N.; Agam, O.; Simon, I.; Balaban, N.Q. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature 2015, 519, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Pearl Mizrahi, S.; Sandler, O.; Lande-Diner, L.; Balaban, N.Q.; Simon, I. Distinguishing between stochasticity and determinism: Examples from cell cycle duration variability. BioEssays News Rev. Mol. Cell. Dev. Biol. 2016, 38, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Yeom, M.; Pendergast, J.S.; Ohmiya, Y.; Yamazaki, S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc. Natl. Acad. Sci. USA 2010, 107, 9665–9670. [Google Scholar] [CrossRef] [PubMed]
- Pendergast, J.S.; Yeom, M.; Reyes, B.A.; Ohmiya, Y.; Yamazaki, S. Disconnected circadian and cell cycles in a tumor-driven cell line. Commun. Integr. Biol. 2010, 3, 536–539. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.B. Diurnal mitotic rhythm in human epidermis. Br. J. Dermatol. 1968, 80, 75–80. [Google Scholar] [CrossRef]
- Cooper, Z.K.; Schiff, A. Mitotic rhythm in human epidermis. Proc. Soc. Exp. Biol. Med. 1938, 39, 323–324. [Google Scholar] [CrossRef]
- Janich, P.; Pascual, G.; Merlos-Suarez, A.; Batlle, E.; Ripperger, J.; Albrecht, U.; Cheng, H.Y.; Obrietan, K.; di Croce, L.; Benitah, S.A. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 2011, 480, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Sporl, F.; Korge, S.; Jurchott, K.; Wunderskirchner, M.; Schellenberg, K.; Heins, S.; Specht, A.; Stoll, C.; Klemz, R.; Maier, B.; et al. Kruppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc. Natl. Acad. Sci. USA 2012, 109, 10903–10908. [Google Scholar] [CrossRef] [PubMed]
- Janich, P.; Toufighi, K.; Solanas, G.; Luis, N.M.; Minkwitz, S.; Serrano, L.; Lehner, B.; Benitah, S.A. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell 2013, 13, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Al-Nuaimi, Y.; Hardman, J.A.; Biro, T.; Haslam, I.S.; Philpott, M.P.; Toth, B.I.; Farjo, N.; Farjo, B.; Baier, G.; Watson, R.E.; et al. A meeting of two chronobiological systems: Circadian proteins period1 and Bmal1 modulate the human hair cycle clock. J. Investig. Dermatol. 2014, 134, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A. Circadian clock-mediated control of stem cell division and differentiation: Beyond night and day. Development 2014, 141, 3105–3111. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Cannon, P.; Mendoza-Viveros, L.; Yuen, A.; Kaern, M.; Cheng, H.Y. The circadian molecular clock regulates adult hippocampal neurogenesis by controlling the timing of cell-cycle entry and exit. Cell Rep. 2013, 5, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.D.; Fentress, S.J.; Qiu, Y.; Yun, K.; Cox, J.S.; Chawla, A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 2013, 341, 1483–1488. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Ferrer, S.; Lucas, D.; Battista, M.; Frenette, P.S. Haematopoietic stem cell release is regulated by circadian oscillations. Nature 2008, 452, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Kettner, N.M. The circadian clock in cancer development and therapy. Prog. Mol. Biol. Transl. Sci. 2013, 119, 221–282. [Google Scholar] [PubMed]
- Davis, S.; Mirick, D.K.; Stevens, R.G. Night shift work, light at night, and risk of breast cancer. J. Natl. Cancer Inst. 2001, 93, 1557–1562. [Google Scholar] [CrossRef] [PubMed]
- Schernhammer, E.S.; Laden, F.; Speizer, F.E.; Willett, W.C.; Hunter, D.J.; Kawachi, I.; Colditz, G.A. Rotating night shifts and risk of breast cancer in women participating in the nurses' health study. J. Natl. Cancer Inst. 2001, 93, 1563–1568. [Google Scholar] [CrossRef] [PubMed]
- Schernhammer, E.S.; Kroenke, C.H.; Laden, F.; Hankinson, S.E. Night work and risk of breast cancer. Epidemiology 2006, 17, 108–111. [Google Scholar] [CrossRef] [PubMed]
- Megdal, S.P.; Kroenke, C.H.; Laden, F.; Pukkala, E.; Schernhammer, E.S. Night work and breast cancer risk: A systematic review and meta-analysis. Eur. J. Cancer 2005, 41, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.L.; Li, L. Association of sleep duration and breast cancer OncotypeDX recurrence score. Breast Cancer Res. Treat. 2012, 134, 1291–1295. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Xiao, Q.; Chu, L.W.; Yu, K.; Matthews, C.E.; Hsing, A.W.; Caporaso, N.E. Sleep duration and cancer in the NIH-AARP diet and health study cohort. PLoS ONE 2016, 11, e0161561. [Google Scholar] [CrossRef] [PubMed]
- Filipski, E.; Innominato, P.F.; Wu, M.; Li, X.M.; Iacobelli, S.; Xian, L.J.; Levi, F. Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl. Cancer Inst. 2005, 97, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Filipski, E.; Li, X.M.; Levi, F. Disruption of circadian coordination and malignant growth. Cancer Causes Control 2006, 17, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Van Dycke, K.C.; Rodenburg, W.; van Oostrom, C.T.; van Kerkhof, L.W.; Pennings, J.L.; Roenneberg, T.; van Steeg, H.; van der Horst, G.T. Chronically alternating light cycles increase breast cancer risk in mice. Curr. Biol. 2015, 25, 1932–1937. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.L.; Luo, H.Y.; Yang, J.; Wu, W.J.; Chen, D.L.; Huang, P.; Xu, R.H. Overexpression of the circadian clock gene bmal1 increases sensitivity to oxaliplatin in colorectal cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014, 20, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.L.; Wu, M.W.; Sun, J.; Sun, Y.L.; Cai, Y.C.; Huang, Y.J.; Xian, L.J. Effects of the biological clock gene Bmal1 on tumour growth and anti-cancer drug activity. J. Biochem. 2010, 148, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.H.; Kim, E.M.; Park, J.K.; Hwang, S.G.; Moon, S.K.; Kim, W.J.; Um, H.D. Bmal1 suppresses cancer cell invasion by blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway. Oncol. Rep. 2013, 29, 2109–2113. [Google Scholar] [PubMed]
- Sakamoto, W.; Takenoshita, S. Overexpression of both clock and Bmal1 inhibits entry to S phase in human colon cancer cells. Fukushima J. Med. Sci. 2015, 61, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J.; Hsieh, A.L.; Sengupta, A.; Krishnanaiah, S.Y.; Stine, Z.E.; Walton, Z.E.; Gouw, A.M.; Venkataraman, A.; Li, B.; Goraksha-Hicks, P.; et al. Myc disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 2015, 22, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Wakabayashi, M.; Hara, Y.; Ishida, N. Tumor growth suppression in vivo by overexpression of the circadian component, per2. Genes Cells Devot. Mol. Cell Mech. 2010, 15, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Hua, H.; Wang, Y.; Wan, C.; Liu, Y.; Zhu, B.; Yang, C.; Wang, X.; Wang, Z.; Cornelissen-Guillaume, G.; Halberg, F. Circadian gene mper2 overexpression induces cancer cell apoptosis. Cancer Sci. 2006, 97, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Katayose, Y.; Yabuuchi, S.; Yamamoto, K.; Mizuma, M.; Shirasou, S.; Onogawa, T.; Ohtsuka, H.; Yoshida, H.; Hayashi, H.; et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 2009, 29, 1201–1209. [Google Scholar] [PubMed]
- Kiessling, S.; Beaulieu-Laroche, L.; Blum, I.D.; Landgraf, D.; Welsh, D.K.; Storch, K.F.; Labrecque, N.; Cermakian, N. Enhancing circadian clock function in cancer cells inhibits tumor growth. BMC Biol. 2017, 15, 13. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Katchy, C.A.; Fu, L. Circadian gene variants in cancer. Ann. Med. 2014, 46, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Reszka, E.; Przybek, M.; Muurlink, O.; Peplonska, B. Circadian gene variants and breast cancer. Cancer Lett. 2017, 390, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Pelicano, H.; Liu, J.; Huang, P.; Lee, C. The circadian gene period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 2002, 111, 41–50. [Google Scholar] [CrossRef]
- Wood, P.A.; Yang, X.; Taber, A.; Oh, E.Y.; Ansell, C.; Ayers, S.E.; Al-Assaad, Z.; Carnevale, K.; Berger, F.G.; Pena, M.M.; et al. Period 2 mutation accelerates ApcMin/+ tumorigenesis. Mol. Cancer Res. 2008, 6, 1786–1793. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Donehower, L.A.; Herron, A.J.; Moore, D.D.; Fu, L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS ONE 2010, 5, e10995. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Bauer, M.R.; Davidson, S.M.; Heimann, M.; Subbaraj, L.; Bhutkar, A.; Bartlebaugh, J.; Vander Heiden, M.G.; Jacks, T. Circadian rhythm disruption promotes lung tumorigenesis. Cell Metab. 2016, 24, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Kettner, N.M.; Voicu, H.; Finegold, M.J.; Coarfa, C.; Sreekumar, A.; Putluri, N.; Katchy, C.A.; Lee, C.; Moore, D.D.; Fu, L. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 2016, 30, 909–924. [Google Scholar] [CrossRef] [PubMed]
- Antoch, M.P.; Toshkov, I.; Kuropatwinski, K.K.; Jackson, M. Deficiency in per proteins has no effect on the rate of spontaneous and radiation-induced carcinogenesis. Cell Cycle 2013, 12, 3673–3680. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in Bmal1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Antoch, M.P.; Gorbacheva, V.Y.; Vykhovanets, O.; Toshkov, I.A.; Kondratov, R.V.; Kondratova, A.A.; Lee, C.; Nikitin, A.Y. Disruption of the circadian clock due to the clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle 2008, 7, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Gauger, M.A.; Sancar, A. Cryptochrome, circadian cycle, cell cycle checkpoints, and cancer. Cancer Res. 2005, 65, 6828–6834. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, N.; Lee, J.H.; Gaddameedhi, S.; Sancar, A. Loss of cryptochrome reduces cancer risk in p53 mutant mice. Proc. Natl. Acad. Sci. USA 2009, 106, 2841–2846. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.H.; McDearmon, E.L.; Panda, S.; Hayes, K.R.; Zhang, J.; Andrews, J.L.; Antoch, M.P.; Walker, J.R.; Esser, K.A.; Hogenesch, J.B.; et al. Circadian and clock-controlled regulation of the mouse transcriptome and cell proliferation. Proc. Natl. Acad. Sci. USA 2007, 104, 3342–3347. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Chen, Y.; Li, X.; Zhao, Q.; Tan, Z. Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem. Funct. 2013, 31, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, B.; Wang, Y.; Sun, N.; Lu, C.; Qian, R.; Hua, L. hClock gene expression in human colorectal carcinoma. Mol. Med. Rep. 2013, 8, 1017–1022. [Google Scholar] [PubMed]
- Karantanos, T.; Theodoropoulos, G.; Gazouli, M.; Vaiopoulou, A.; Karantanou, C.; Lymberi, M.; Pektasides, D. Expression of clock genes in patients with colorectal cancer. Int. J. Biol. Markers 2013, 28, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Takenoshita, S.; Akaike, M.; Kunisaki, C.; Fujii, S.; Nozaki, A.; Numata, K.; Shiozawa, M.; Rino, Y.; Tanaka, K.; et al. Expression of circadian genes correlates with liver metastasis and outcomes in colorectal cancer. Oncol. Rep. 2011, 25, 1439–1446. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, R.; Sun, N.; Lu, C.; Chen, Z.; Hua, L. Circadian gene hClock enhances proliferation and inhibits apoptosis of human colorectal carcinoma cells in vitro and in vivo. Mol. Med. Rep. 2015, 11, 4204–4210. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Chang, A.K.; Zang, M.X.; Bi, H.; Li, S.; Wang, M.; Xing, X.; Wu, H. Induction of the clock gene by e2-eralpha signaling promotes the proliferation of breast cancer cells. PLoS ONE 2014, 9, e95878. [Google Scholar]
- Roe, O.D.; Anderssen, E.; Helge, E.; Pettersen, C.H.; Olsen, K.S.; Sandeck, H.; Haaverstad, R.; Lundgren, S.; Larsson, E. Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: The emerging gene portrait of the mesothelioma phenotype. PLoS ONE 2009, 4, e6554. [Google Scholar] [CrossRef] [PubMed]
- Elshazley, M.; Sato, M.; Hase, T.; Yamashita, R.; Yoshida, K.; Toyokuni, S.; Ishiguro, F.; Osada, H.; Sekido, Y.; Yokoi, K.; et al. The circadian clock gene Bmal1 is a novel therapeutic target for malignant pleural mesothelioma. Int. J. Cancer 2012, 131, 2820–2831. [Google Scholar] [CrossRef] [PubMed]
- Puram, R.V.; Kowalczyk, M.S.; de Boer, C.G.; Schneider, R.K.; Miller, P.G.; McConkey, M.; Tothova, Z.; Tejero, H.; Heckl, D.; Jaras, M.; et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell 2016, 165, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Altman, B.J. Cancer clocks out for lunch: Disruption of circadian rhythm and metabolic oscillation in cancer. Front. Cell Dev. Biol. 2016, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Relogio, A.; Thomas, P.; Medina-Perez, P.; Reischl, S.; Bervoets, S.; Gloc, E.; Riemer, P.; Mang-Fatehi, S.; Maier, B.; Schafer, R.; et al. Ras-mediated deregulation of the circadian clock in cancer. PLoS Genet. 2014, 10, e1004338. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. Myc on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Shostak, A.; Ruppert, B.; Ha, N.; Bruns, P.; Toprak, U.H.; Project, I.M.-S.; Eils, R.; Schlesner, M.; Diernfellner, A.; Brunner, M. Myc/Miz1-dependent gene repression inversely coordinates the circadian clock with cell cycle and proliferation. Nat. Commun. 2016, 7, 11807. [Google Scholar] [CrossRef] [PubMed]
- Shostak, A.; Diernfellner, A.; Brunner, M. Myc inhibits the clock and supports proliferation. Cell Cycle 2016, 15, 3323–3324. [Google Scholar] [CrossRef] [PubMed]
- Michael, A.K.; Harvey, S.L.; Sammons, P.J.; Anderson, A.P.; Kopalle, H.M.; Banham, A.H.; Partch, C.L. Cancer/testis antigen pasd1 silences the circadian clock. Mol. Cell 2015, 58, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Huisman, S.A.; Oklejewicz, M.; Ahmadi, A.R.; Tamanini, F.; Ijzermans, J.N.; van der Horst, G.T.; de Bruin, R.W. Colorectal liver metastases with a disrupted circadian rhythm phase shift the peripheral clock in liver and kidney. Int. J. Cancer 2015, 136, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Masri, S.; Papagiannakopoulos, T.; Kinouchi, K.; Liu, Y.; Cervantes, M.; Baldi, P.; Jacks, T.; Sassone-Corsi, P. Lung adenocarcinoma distally rewires hepatic circadian homeostasis. Cell 2016, 165, 896–909. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Son, G.H.; Kim, K. Circadian rhythm of adrenal glucocorticoid: Its regulation and clinical implications. Biochim. Biophys. Acta 2011, 1812, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Zelinka, T.; Strauch, B.; Pecen, L.; Widimsky, J., Jr. Diurnal blood pressure variation in pheochromocytoma, primary aldosteronism and cushing’s syndrome. J. Hum. Hypertens. 2004, 18, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Zelinka, T.; Widimsky, J.; Weisserova, J. Diminished circadian blood pressure rhythm in patients with asymptomatic normotensive pheochromocytoma. Physiol. Res. 2001, 50, 631–634. [Google Scholar] [PubMed]
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shostak, A. Circadian Clock, Cell Division, and Cancer: From Molecules to Organism. Int. J. Mol. Sci. 2017, 18, 873. https://doi.org/10.3390/ijms18040873
Shostak A. Circadian Clock, Cell Division, and Cancer: From Molecules to Organism. International Journal of Molecular Sciences. 2017; 18(4):873. https://doi.org/10.3390/ijms18040873
Chicago/Turabian StyleShostak, Anton. 2017. "Circadian Clock, Cell Division, and Cancer: From Molecules to Organism" International Journal of Molecular Sciences 18, no. 4: 873. https://doi.org/10.3390/ijms18040873