Circadian Rhythm Neuropeptides in Drosophila: Signals for Normal Circadian Function and Circadian Neurodegenerative Disease
Abstract
:1. Introduction
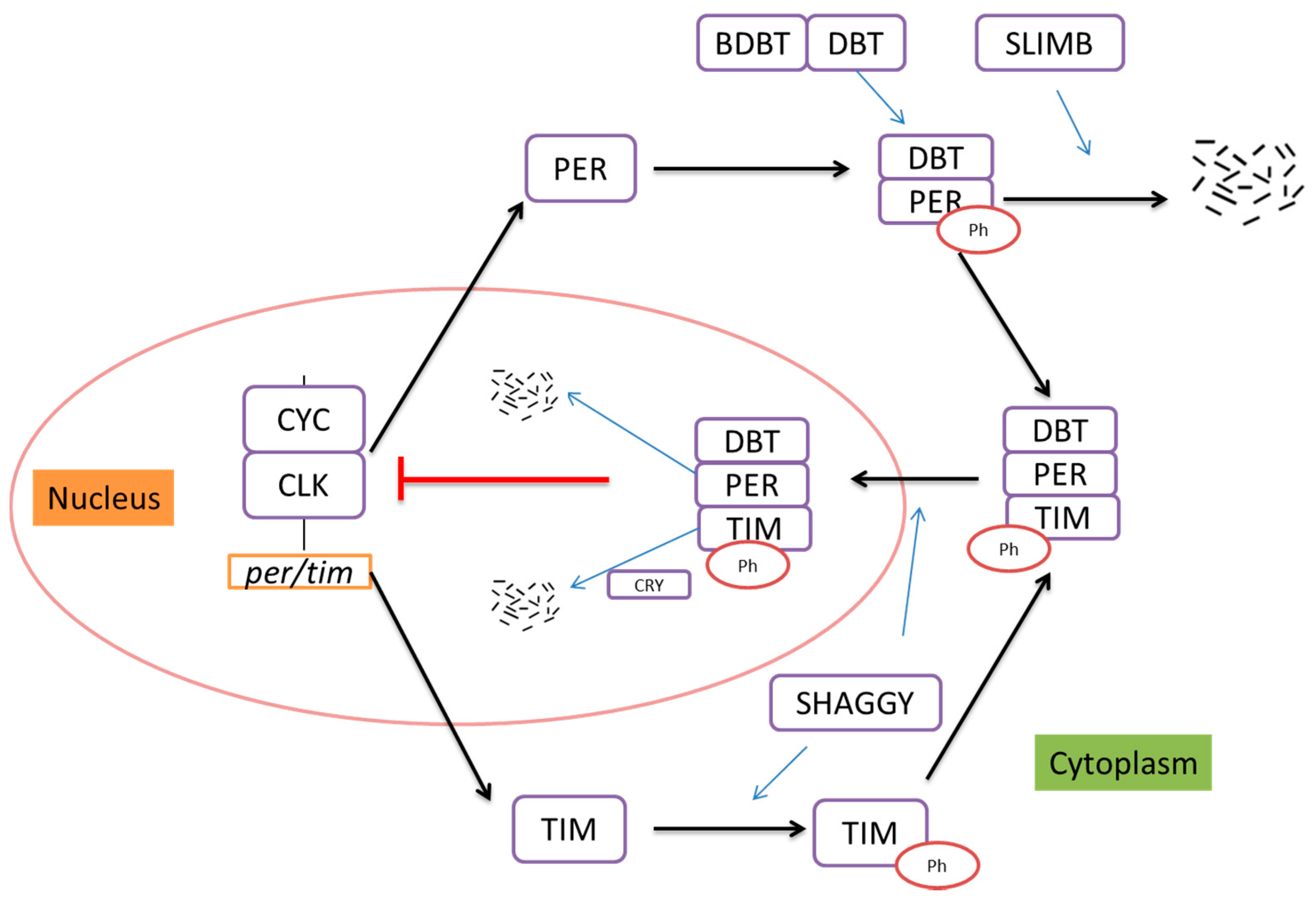
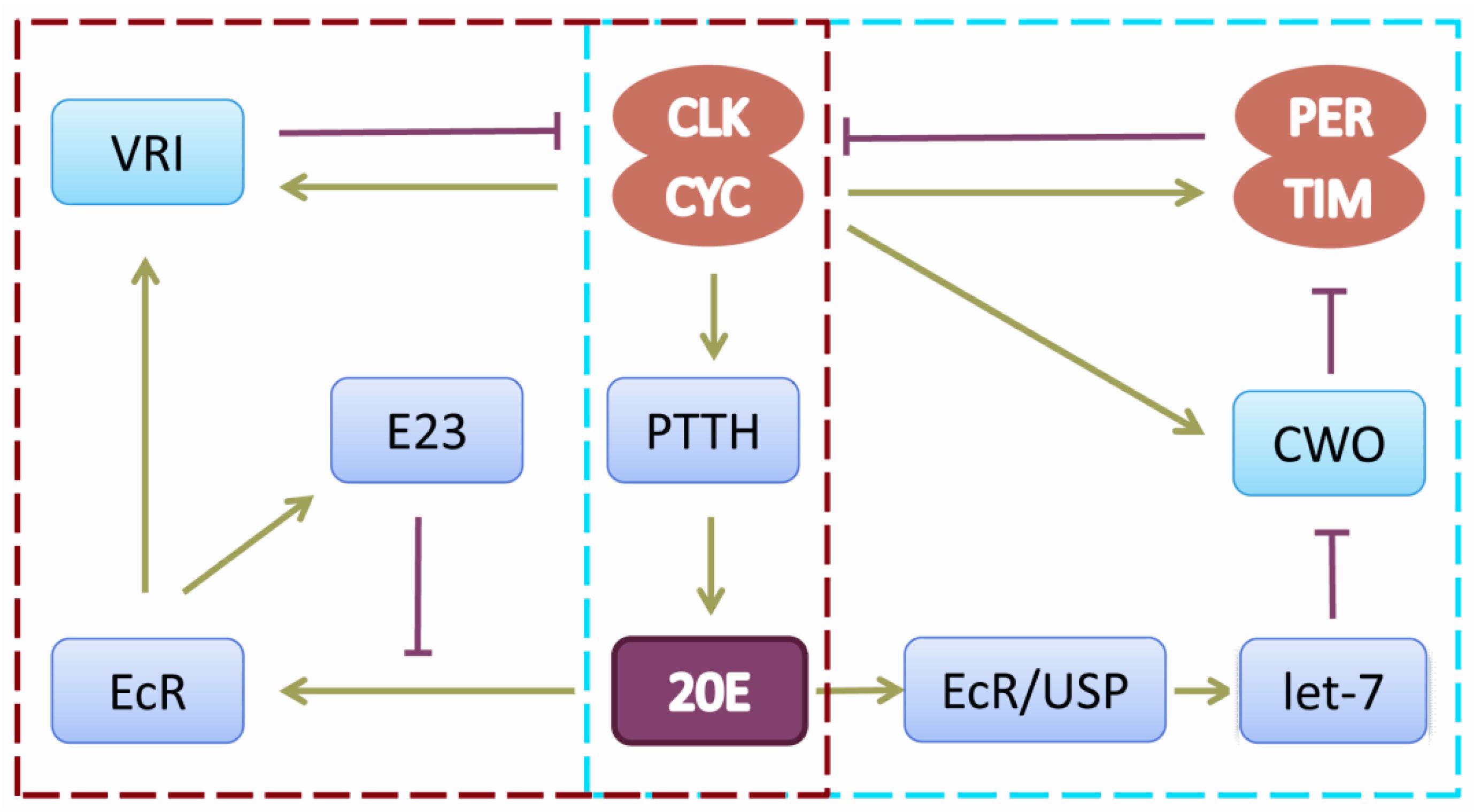
2. The Drosophila Circadian Rhythm and Neuroendocrine Regulation
3. Circadian Rhythms, Neuropeptides and Neurodegenerative Disease in Drosophila
4. Perspectives
Acknowledgments
Conflicts of Interest
References
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Konopka, R.J.; Benzer, S. Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1971, 68, 2112–2116. [Google Scholar] [CrossRef] [PubMed]
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Lin, M.C.; Glossop, N.R.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Blau, J. Vrille, PDP1, and dclock form a second feedback loop in the Drosophila circadian clock. Cell 2003, 112, 329–341. [Google Scholar] [CrossRef]
- Sehgal, A.; Price, J.L.; Man, B.; Young, M.W. Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 1994, 263, 1603–1606. [Google Scholar] [CrossRef] [PubMed]
- Reppert, S.M.; Sauman, I. Period and timeless tango: A dance of two clock genes. Neuron 1995, 15, 983–986. [Google Scholar] [CrossRef]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef]
- Fan, J.Y.; Agyekum, B.; Venkatesan, A.; Hall, D.R.; Keightley, A.; Bjes, E.S.; Bouyain, S.; Price, J.L. Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron 2013, 80, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Blau, J.; Young, M.W. Cycling vrille expression is required for a functional Drosophila clock. Cell 1999, 99, 661–671. [Google Scholar] [CrossRef]
- Rutila, J.E.; Suri, V.; Le, M.; So, W.V.; Rosbash, M.; Hall, J.C. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of and transcription of Drosophila period and timeless. Cell 1998, 93, 805–814. [Google Scholar] [CrossRef]
- Emery, P.; Al, E. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef]
- Martinek, S.; Inonog, S.; Manoukian, A.S.; Young, M.W. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 2001, 105, 769. [Google Scholar] [CrossRef]
- Allada, R.; White, N.E.; So, W.V.; Hall, J.C.; Rosbash, M. A mutant Drosophila homolog of mammalian clock disrupts circadian rhythms and transcription of period and timeless. Cell 1998, 93, 791–804. [Google Scholar] [CrossRef]
- Glossop, N.R.; Houl, J.H.; Zheng, H.; Ng, F.S.; Dudek, S.M.; Hardin, P.E. Vrille feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 2003, 37, 249–261. [Google Scholar] [CrossRef]
- Chiu, J.C.; Ko, H.W.; Edery, I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 2011, 145, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Houl, J.H.; Hardin, P.E. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr. Biol. 2011, 21, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.M.; Kilman, V.L.; Keegan, K.; Paddock, B.; Emery-Le, M.; Rosbash, M.; Allada, R. A role for casein kinase 2α in the Drosophila circadian clock. Nature 2002, 420, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Panda, S.; Hogenesch, J.B.; Kay, S.A. Circadian rhythms from flies to human. Nature 2002, 417, 329. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E. The circadian timekeeping system of Drosophila. Curr. Biol. 2005, 15, R714–R722. [Google Scholar] [CrossRef] [PubMed]
- Peschel, N.; Helfrich-Forster, C. Setting the clock—By nature: Circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 2011, 585, 1435–1442. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E.; Panda, S. Circadian timekeeping and output mechanisms in animals. Curr. Opin. Neurobiol. 2013, 23, 724–731. [Google Scholar] [CrossRef] [PubMed]
- Rivas, G.B.; Bauzer, L.G.; Meireles-Filho, A.C. The environment is everything that isn’t me: Molecular mechanisms and evolutionary dynamics of insect clocks in variable surroundings. Front. Physiol. 2015, 6, 400. [Google Scholar] [CrossRef] [PubMed]
- Darlington, T.K.; Wagersmith, K.; Ceriani, M.F.; Staknis, D.; Gekakis, N.; Steeves, T.D.L.; Weitz, C.J.; Takahashi, J.S.; Kay, S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 1998, 280, 1599–1603. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.W.; Jiang, J.; Edery, I. Role for slimb in the degradation of Drosophila period protein phosphorylated by doubletime. Nature 2002, 420, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Rothenfluh, A.; Young, M.W.; Saez, L. A timeless-independent function for period proteins in the Drosophila clock. Neuron 2000, 26, 505. [Google Scholar] [CrossRef]
- Lee, C.; Bae, K.; Edery, I. PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: A basis for circadian transcription. Mol. Cell. Biol. 1999, 19, 5316–5325. [Google Scholar] [CrossRef] [PubMed]
- Jaumouille, E.; Machado Almeida, P.; Stahli, P.; Koch, R.; Nagoshi, E. Transcriptional regulation via nuclear receptor crosstalk required for the Drosophila circadian clock. Curr. Biol. 2015, 25, 1502–1508. [Google Scholar] [CrossRef] [PubMed]
- Richier, B.; Michard-Vanhee, C.; Lamouroux, A.; Papin, C.; Rouyer, F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J. Biol. Rhythms 2008, 23, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, W.; Hardin, P.E. CLOCKWORK ORANGE enhances PERIOD mediated rhythms in transcriptional repression by antagonizing E-box binding by CLOCK-CYCLE. PLoS Genet. 2016, 12, e1006430. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; McNeil, G.P.; Jackson, F.R. Translational regulation of the doubletime/ckidelta/epsilon kinase by lark contributes to circadian period modulation. PLoS Genet. 2014, 10, e1004536. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Lee, J.; Choi, C.; Kilman, V.L.; Kim, J.; Park, S.M.; Jang, S.K.; Allada, R.; Choe, J. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 2011, 470, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Allada, R. Ataxin-2 activates period translation to sustain circadian rhythms in Drosophila. Science 2013, 340, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Challet, E. Keeping circadian time with hormones. Diabetes Obes. Metab. 2015, 17, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Grima, B.; Chelot, E.; Xia, R.; Rouyer, F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 2004, 431, 869–873. [Google Scholar] [CrossRef] [PubMed]
- Stoleru, D.; Peng, Y.; Agosto, J.; Rosbash, M. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 2004, 431, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Bloch, G.; Hazan, E.; Rafaeli, A. Circadian rhythms and endocrine functions in adult insects. J. Insect Physiol. 2013, 59, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Helfrich-FÖRster, C. Neuropeptide PDF plays multiple roles in the circadian clock of Drosophila melanogaster. Sleep Biol. Rhythms 2009, 7, 130–143. [Google Scholar] [CrossRef]
- Guo, F.; Cerullo, I.; Chen, X.; Rosbash, M. PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife Sci. 2014, 3, e02780. [Google Scholar]
- Klose, M.; Duvall, L.B.; Li, W.; Liang, X.; Ren, C.; Steinbach, J.H.; Taghert, P.H. Functional PDF signaling in the Drosophila circadian neural circuit is gated by Ral A-dependent modulation. Neuron 2016, 90, 781–794. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, F.; Shen, J.; Rosbash, M. PDF and camp enhance per stability in Drosophila clock neurons. Proc. Natl. Acad. Sci. USA 2014, 111, 1284–1290. [Google Scholar] [CrossRef] [PubMed]
- Tomioka, K.; Miyasako, Y.; Umezaki, Y. PDF as a coupling mediator between the light-entrainable and temperature-entrainable clocks in Drosophila melanogaster. Acta Biol. Hung. 2008, 59, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Seluzicki, A.; Flourakis, M.; Kula-Eversole, E.; Zhang, L.; Kilman, V.; Allada, R. Dual PDF signaling pathways reset clocks via timeless and acutely excite target neurons to control circadian behavior. PLoS Biol. 2014, 12, e1001810. [Google Scholar] [CrossRef] [PubMed]
- Parisky, K.M.; Agosto, J.; Pulver, S.R.; Shang, Y.; Kuklin, E.; Hodge, J.J.; Kang, K.; Liu, X.; Garrity, P.A.; Rosbash, M.; et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 2008, 60, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Renn, S.C.; Park, J.H.; Rosbash, M.; Hall, J.C.; Taghert, P.H. A PDF neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 1999, 99, 791–802. [Google Scholar] [CrossRef]
- Chung, B.Y.; Kilman, V.L.; Keath, J.R.; Pitman, J.L.; Allada, R. The GABA A receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 2009, 19, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Sheeba, V.; Fogle, K.J.; Kaneko, M.; Rashid, S.; Chou, Y.T.; Sharma, V.K.; Holmes, T.C. Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 2008, 18, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Stormo, G.D.; Taghert, P.H. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J. Neurosci. 2004, 24, 7951–7957. [Google Scholar] [CrossRef] [PubMed]
- Taghert, P.H.; Nitabach, M.N. Peptide neuromodulation in invertebrate model systems. Neuron 2012, 76, 82–97. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Cong, X.; Zhang, R.; Wu, D.; An, C.; Zhao, Z. Regulation of circadian locomotor rhythm by neuropeptide Y-like system in Drosophila melanogaster. Insect Mol. Biol. 2013, 22, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Johard, H.A.; Yoishii, T.; Dircksen, H.; Cusumano, P.; Rouyer, F.; Helfrich-Forster, C.; Nassel, D.R. Peptidergic clock neurons in Drosophila: Ion transport peptide and short neuropeptide f in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 2009, 516, 59–73. [Google Scholar] [CrossRef] [PubMed]
- Dubruille, R.; Emery, P. A plastic clock: How circadian rhythms respond to environmental cues in Drosophila. Mol. Neurobiol. 2008, 38, 129–145. [Google Scholar] [CrossRef] [PubMed]
- Hermann, C.; Yoshii, T.; Dusik, V.; Helfrich-Forster, C. Neuropeptide F immunoreactive clock neurons modify evening locomotor activity and free-running period in Drosophila melanogaster. J. Comp. Neurol. 2012, 520, 970–987. [Google Scholar] [CrossRef] [PubMed]
- Erion, R.; King, A.N.; Wu, G.; Hogenesch, J.B.; Sehgal, A. Neural clocks and neuropeptide F/Y regulate circadian gene expression in a peripheral metabolic tissue. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Shi, W.; Li, L.; Zheng, Z.; Li, T.; Bai, W.; Zhao, Z. Regulation of sleep by the short neuropeptide F (SNPF) in Drosophila melanogaster. Insect Biochem. Mol. Biol. 2013, 43, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Donelson, N.C.; Vecsey, C.G.; Guo, F.; Rosbash, M.; Griffith, L.C. Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 2013, 80, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Shafer, O.T. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 2014, 343, 1516–1520. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Wang, H.; Liu, Z.; He, C.; An, C.; Zhao, Z. Regulation of sleep by insulin-like peptide system in Drosophila melanogaster. Sleep 2015, 38, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Barber, A.F.; Erion, R.; Holmes, T.C.; Sehgal, A. Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 2016, 30, 2596–2606. [Google Scholar] [CrossRef] [PubMed]
- Monyak, R.E.; Emerson, D.; Schoenfeld, B.P.; Zheng, X.; Chambers, D.B.; Rosenfelt, C.; Langer, S.; Hinchey, P.; Choi, C.H.; McDonald, T.V.; et al. Insulin signaling misregulation underlies circadian and cognitive deficits in a Drosophila fragile X model. Mol. Psychiatry 2016. [Google Scholar] [CrossRef] [PubMed]
- Hermann-Luibl, C.; Yoshii, T.; Senthilan, P.R.; Dircksen, H.; Helfrich-Forster, C. The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J. Neurosci. 2014, 34, 9522–9536. [Google Scholar] [CrossRef] [PubMed]
- Kunst, M.; Hughes, M.E.; Raccuglia, D.; Felix, M.; Li, M.; Barnett, G.; Duah, J.; Nitabach, M.N. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 2014, 24, 2652–2664. [Google Scholar] [CrossRef] [PubMed]
- Goda, T.; Tang, X.; Umezaki, Y.; Chu, M.L.; Hamada, F.N. Drosophila DH31 neuropeptide and PDF receptor regulate night-onset temperature preference. J. Neurosci. 2016, 36, 11739–11754. [Google Scholar] [CrossRef] [PubMed]
- Cavey, M.; Collins, B.; Bertet, C.; Blau, J. Circadian rhythms in neuronal activity propagate through output circuits. Nat. Neurosci. 2016, 19, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Cavanaugh, D.J.; Geratowski, J.D.; Wooltorton, J.R.; Spaethling, J.M.; Hector, C.E.; Zheng, X.; Johnson, E.C.; Eberwine, J.H.; Sehgal, A. Identification of a circadian output circuit for rest: Activity rhythms in Drosophila. Cell 2014, 157, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Reiher, W.; Hermann-Luibl, C.; Sellami, A.; Cognigni, P.; Kondo, S.; Helfrich-Förster, C.; Veenstra, J.A.; Wegener, C. Allatostatin a signalling in Drosophila regulates feeding and sleep and is modulated by pdf. PLoS Genet. 2016, 12, e1006346. [Google Scholar]
- Nassel, D.R.; Winther, A.M. Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 2010, 92, 42–104. [Google Scholar] [CrossRef] [PubMed]
- Selbie, L.A.; Hill, S.J. G protein-coupled-receptor cross-talk: The fine-tuning of multiple receptor-signalling pathways. Science 1998, 19, 87–93. [Google Scholar] [CrossRef]
- Majercak, J.; Kalderon, D.; Edery, I. Drosophila melanogaster deficient in protein kinase a manifests behavior-specific arrhythmia but normal clock function. Mol. Cell. Biol. 1997, 17, 5915–5922. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Sedore, S.A.; Cronmiller, C.; Hirsh, J. Type II camp-dependent protein kinase-deficient Drosophila are viable but show developmental, circadian, and drug response phenotypes. J. Biol. Chem. 2000, 275, 20588–20596. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, E.K. AMP-activated protein kinase as a key molecular link between metabolism and clockwork. Exp. Mol. Med. 2013, 45, e33. [Google Scholar] [CrossRef] [PubMed]
- Ishimoto, H.; Sakai, T.; Kitamoto, T. Ecdysone signaling regulates the formation of long-term courtship memory in adult Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2009, 106, 6381–6386. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, Z.; Li, T.; Zhang, R.; Xue, Y.; Zhong, Y.; Bai, W.; Zhou, D.; Zhao, Z. Regulation of Drosophila circadian rhythms by miRNA Let-7 is mediated by a regulatory cycle. Nat. Commun. 2014, 5, 5549. [Google Scholar] [CrossRef] [PubMed]
- McBrayer, Z.; Ono, H.; Shimell, M.; Parvy, J.P.; Beckstead, R.B.; Warren, J.T.; Thummel, C.S.; Dauphin-Villemant, C.; Gilbert, L.I.; O’Connor, M.B. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 2007, 13, 857–871. [Google Scholar] [CrossRef] [PubMed]
- Gauhar, Z.; Sun, L.V.; Hua, S.; Mason, C.E.; Fuchs, F.; Li, T.R.; Boutros, M.; White, K.P. Genomic mapping of binding regions for the ecdysone receptor protein complex. Genome Res. 2009, 19, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.Q.; Tanimura, T.; Matsumoto, A. Membrane-bound transporter controls the circadian transcription of clock genes in Drosophila. Genes Cells 2011, 16, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Di Cara, F.; King-Jones, K. The circadian clock is a key driver of steroid hormone production in Drosophila. Curr. Biol. 2016, 26, 2469–2477. [Google Scholar] [CrossRef] [PubMed]
- Hamasaka, Y.; Nassel, D.R. Mapping of serotonin, dopamine, and histamine in relation to different clock neurons in the brain of Drosophila. J. Comp. Neurol. 2006, 494, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Ng, F.; Lebestky, T.; Grygoruk, A.; Djapri, C.; Lawal, H.O.; Zaveri, H.A.; Mehanzel, F.; Najibi, R.; Seidman, G. Dispensable, redundant, complementary, and cooperative roles of dopamine, octopamine, and serotonin in Drosophila melanogaster. Genetics 2013, 193, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, M.S.; Tempel, B.L. Genetic dissection of monoamine neurotransmitter synthesis in Drosophila. Nature 1983, 303, 67. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Lin, F.; Zheng, X.; Sehgal, A. Serotonin modulates circadian entrainment in Drosophila. Neuron 2005, 47, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Balfanz, S.; Strunker, T.; Frings, S.; Baumann, A. A family of octopamine receptors that specifically induce cyclic AMP production or Ca2+ release in Drosophila melanogaster. J. Neurochem. 2005, 93, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Andretic, R.; Hirsh, J. Circadian modulation of dopamine receptor responsiveness in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 1873. [Google Scholar] [CrossRef] [PubMed]
- Nall, A.; Sehgal, A. Monoamines and sleep in Drosophila. Behav. Neurosci. 2014, 128, 264. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Kume, S.; Park, S.K.; Hirsh, J.; Jackson, F.R. Dopamine is a regulator of arousal in the fruit fly. J. Neurosci. 2005, 25, 7377–7384. [Google Scholar] [CrossRef] [PubMed]
- Crocker, A.; Sehgal, A. Octopamine regulates sleep in Drosophila through protein kinase A-dependent mechanisms. J. Neurosci. 2008, 28, 9377–9385. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Joiner, W.J.; Sehgal, A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 2006, 16, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Lelito, K.R.; Shafer, O.T. Reciprocal cholinergic and gabaergic modulation of the small ventrolateral pacemaker neurons of Drosophila’s circadian clock neuron network. J. Neurophysiol. 2012, 107, 2096–2108. [Google Scholar] [CrossRef] [PubMed]
- Hamasaka, Y.; Rieger, D.; Parmentier, M.L.; Grau, Y.; Helfrich-Forster, C.; Nassel, D.R. Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J. Comp. Neurol. 2007, 505, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.; Kaplan, H.S.; Cavey, M.; Lelito, K.R.; Bahle, A.H.; Zhu, Z.; Macara, A.M.; Roman, G.; Shafer, O.T.; Blau, J. Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol. 2014, 12, e1001959. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.; Kane, E.A.; Reeves, D.C.; Akabas, M.H.; Blau, J. Balance of activity between LNvs and glutamatergic dorsal clock neurons promotes robust circadian rhythms in Drosophila. Neuron 2012, 74, 706–718. [Google Scholar] [CrossRef] [PubMed]
- Dahdal, D.; Reeves, D.C.; Ruben, M.; Akabas, M.H.; Blau, J. Drosophila pacemaker neurons require g protein signaling and gabaergic inputs to generate twenty-four hour behavioral rhythms. Neuron 2010, 68, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Hamasaka, Y.; Wegener, C.; Nassel, D.R. GABA modulates Drosophila circadian clock neurons via GABAB receptors and decreases in calcium. J. Neurobiol. 2005, 65, 225–240. [Google Scholar] [CrossRef] [PubMed]
- Mattis, J.; Sehgal, A. Circadian rhythms, sleep, and disorders of aging. Trends Endocrinol. Metab. 2016, 27, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.E.; McLeland, J.S.; Toedebusch, C.D.; Xiong, C.; Fagan, A.M.; Duntley, S.P.; Morris, J.C.; Holtzman, D.M. Sleep quality and preclinical Alzheimer disease. JAMA Neurol. 2013, 70, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Blake, M.R.; Holbrook, S.D.; Kotwica-Rolinska, J.; Chow, E.S.; Kretzschmar, D.; Giebultowicz, J.M. Manipulations of amyloid precursor protein cleavage disrupt the circadian clock in aging Drosophila. Neurobiol. Dis. 2015, 77, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Long, D.M.; Blake, M.R.; Dutta, S.; Holbrook, S.D.; Kotwica-Rolinska, J.; Kretzschmar, D.; Giebultowicz, J.M. Relationships between the circadian system and Alzheimer’s disease-like symptoms in Drosophila. PLoS ONE 2014, 9, e106068. [Google Scholar] [CrossRef] [PubMed]
- Kolker, D.E.; Fukuyama, H.; Huang, D.S.; Takahashi, J.S.; Horton, T.H.; Turek, F.W. Aging alters circadian and light-induced expression of clock genes in golden hamsters. J. Biol. Rhythms 2003, 18, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Wyse, C.A.; Coogan, A.N. Impact of aging on diurnal expression patterns of clock and bmal1 in the mouse brain. Brain Res. 2010, 1337, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.C.; Guarente, L. Sirt1 mediates central circadian control in the SCN by a mechanism that decays with aging. Cell 2013, 153, 1448–1460. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Straume, M.; Tei, H.; Sakaki, Y.; Menaker, M.; Block, G.D. Effects of aging on central and peripheral mammalian clocks. Proc. Natl. Acad. Sci. USA 2002, 99, 10801–10806. [Google Scholar] [CrossRef] [PubMed]
- Roozendaal, B.; van Gool, W.A.; Swaab, D.F.; Hoogendijk, J.E.; Mirmiran, M. Changes in vasopressin cells of the rat suprachiasmatic nucleus with aging. Brain Res. 1987, 409, 259–264. [Google Scholar] [CrossRef]
- Nygard, M.; Hill, R.H.; Wikstrom, M.A.; Kristensson, K. Age-related changes in electrophysiological properties of the mouse suprachiasmatic nucleus in vitro. Brain Res. Bull. 2005, 65, 149–154. [Google Scholar] [CrossRef] [PubMed]
- Farajnia, S.; Michel, S.; Deboer, T.; vanderLeest, H.T.; Houben, T.; Rohling, J.H.; Ramkisoensing, A.; Yasenkov, R.; Meijer, J.H. Evidence for neuronal desynchrony in the aged suprachiasmatic nucleus clock. J. Neurosci. 2012, 32, 5891–5899. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Chen, W.F.; Yue, Z.; Chen, D.; Sowcik, M.; Sehgal, A.; Zheng, X. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell 2012, 11, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.; Evans, J.M.; Hendricks, J.C.; Sehgal, A. A Drosophila model for age-associated changes in sleep: Wake cycles. Proc. Natl. Acad. Sci. USA 2006, 103, 13843–13847. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, K.; Krishnan, N.; Guzik, E.M.; Pyza, E.; Giebultowicz, J.M. Effects of aging on the molecular circadian oscillations in Drosophila. Chronobiol. Int. 2012, 29, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Rakshit, K.; Giebultowicz, J.M. Cryptochrome restores dampened circadian rhythms and promotes healthspan in aging Drosophila. Aging Cell 2013, 12, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Umezaki, Y.; Yoshii, T.; Kawaguchi, T.; Helfrich-Forster, C.; Tomioka, K. Pigment-dispersing factor is involved in age-dependent rhythm changes in Drosophila melanogaster. J. Biol. Rhythms 2012, 27, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.F.; Possidente, B.; Lomas, D.A.; Crowther, D.C. The central molecular clock is robust in the face of behavioural arrhythmia in a Drosophila model of Alzheimer’s disease. Dis. Model. Mech. 2014, 7, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Means, J.C.; Venkatesan, A.; Gerdes, B.; Fan, J.Y.; Bjes, E.S.; Price, J.L. Drosophila spaghetti and doubletime link the circadian clock and light to caspases, apoptosis and tauopathy. PLoS Genet. 2015, 11, e1005171. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Y.; Means, J.C.; Bjes, E.S.; Price, J.L. Drosophila DBT autophosphorylation of its C-terminal domain antagonized by SPAG and involved in UV-induced apoptosis. Mol. Cell. Biol. 2015, 35, 2414–2424. [Google Scholar] [CrossRef] [PubMed]
- De Calignon, A.; Fox, L.M.; Pitstick, R.; Carlson, G.A.; Bacskai, B.J.; Spires-Jones, T.L.; Hyman, B.T. Caspase activation precedes and leads to tangles. Nature 2010, 464, 1201–1204. [Google Scholar] [CrossRef] [PubMed]
- Rohn, T.T.; Rissman, R.A.; Davis, M.C.; Kim, Y.E.; Cotman, C.W.; Head, E. Caspase 9 activation and caspase cleavage of tau in the Alzheimer’s disease brain. Neurobiol. Dis. 2002, 11, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, K.I.; Cantera, R. Circadian rhythms in the morphology of neurons in Drosophila. Cell Tissue Res. 2011, 344, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Schoenmann, Z.; Assa-Kunik, E.; Tiomny, S.; Minis, A.; Haklai-Topper, L.; Arama, E.; Yaron, A. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J. Neurosci. 2010, 30, 6375–6386. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Vykhovanets, O.; Kondratova, A.A.; Antoch, M.P. Antioxidant N-acetyl-l-cysteine ameliorates symptoms of premature aging associated with the deficiency of the circadian protein bmal1. Aging (Albany NY) 2009, 1, 979–987. [Google Scholar] [CrossRef] [PubMed]
- Musiek, E.S.; Lim, M.M.; Yang, G.; Bauer, A.Q.; Qi, L.; Lee, Y.; Roh, J.H.; Ortiz-Gonzalez, X.; Dearborn, J.T.; Culver, J.P.; et al. Circadian clock proteins regulate neuronal redox homeostasis and neurodegeneration. J. Clin. Investig. 2013, 123, 5389–5400. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Kretzschmar, D.; Rakshit, K.; Chow, E.; Giebultowicz, J.M. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging (Albany NY) 2009, 1, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Rakshit, K.; Chow, E.S.; Wentzell, J.S.; Kretzschmar, D.; Giebultowicz, J.M. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiol. Dis. 2012, 45, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Kuintzle, R.C.; Chow, E.S.; Westby, T.N.; Gvakharia, B.O.; Giebultowicz, J.M.; Hendrix, D.A. Circadian deep sequencing reveals stress-response genes that adopt robust rhythmic expression during aging. Nat. Commun. 2017, 8, 14529. [Google Scholar] [CrossRef] [PubMed]
- Vaccaro, A.; Issa, A.R.; Seugnet, L.; Birman, S.; Klarsfeld, A. Drosophila clock is required in brain pacemaker neurons to prevent premature locomotor aging independently of its circadian function. PLoS Genet. 2017, 13, e1006507. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Sehgal, A. AKT and TOR signaling set the pace of the circadian pacemaker. Curr. Biol. 2010, 20, 1203–1208. [Google Scholar] [CrossRef] [PubMed]
- Metaxakis, A.; Tain, L.S.; Gronke, S.; Hendrich, O.; Hinze, Y.; Birras, U.; Partridge, L. Lowered insulin signalling ameliorates age-related sleep fragmentation in Drosophila. PLoS Biol. 2014, 12, e1001824. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ling, J.; Yuan, C.; Dubruille, R.; Emery, P. A role for Drosophila ATX2 in activation of per translation and circadian behavior. Science 2013, 340, 879–882. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Turm, H.; Bekenstein, U.; Shifman, S.; Kadener, S. A new in vivo model of pantothenate kinase-associated neurodegeneration reveals a surprising role for transcriptional regulation in pathogenesis. Front. Cell Neurosci. 2013, 7, 146. [Google Scholar] [PubMed]
- Gekakis, N.; Staknis, D.; Nguyen, H.B.; Davis, F.C.; Wilsbacher, L.D.; King, D.P.; Takahashi, J.S.; Weitz, C.J. Role of the clock protein in the mammalian circadian mechanism. Science 1998, 280, 1564–1569. [Google Scholar] [CrossRef] [PubMed]
- Kume, K.; Zylka, M.J.; Sriram, S.; Shearman, L.P.; Weaver, D.R.; Jin, X.; Maywood, E.S.; Hastings, M.H.; Reppert, S.M. MCRY1 and MCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 1999, 98, 193. [Google Scholar] [CrossRef]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Q.; Wu, B.; Price, J.L.; Zhao, Z. Circadian Rhythm Neuropeptides in Drosophila: Signals for Normal Circadian Function and Circadian Neurodegenerative Disease. Int. J. Mol. Sci. 2017, 18, 886. https://doi.org/10.3390/ijms18040886
He Q, Wu B, Price JL, Zhao Z. Circadian Rhythm Neuropeptides in Drosophila: Signals for Normal Circadian Function and Circadian Neurodegenerative Disease. International Journal of Molecular Sciences. 2017; 18(4):886. https://doi.org/10.3390/ijms18040886
Chicago/Turabian StyleHe, Qiankun, Binbin Wu, Jeffrey L. Price, and Zhangwu Zhao. 2017. "Circadian Rhythm Neuropeptides in Drosophila: Signals for Normal Circadian Function and Circadian Neurodegenerative Disease" International Journal of Molecular Sciences 18, no. 4: 886. https://doi.org/10.3390/ijms18040886






