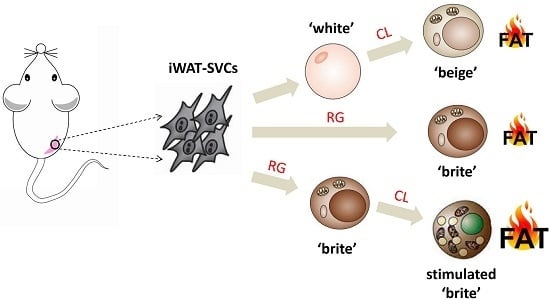
An Additive Effect of Promoting Thermogenic Gene Expression in Mice Adipose-Derived Stromal Vascular Cells by Combination of Rosiglitazone and CL316,243
Abstract
:1. Introduction
2. Results
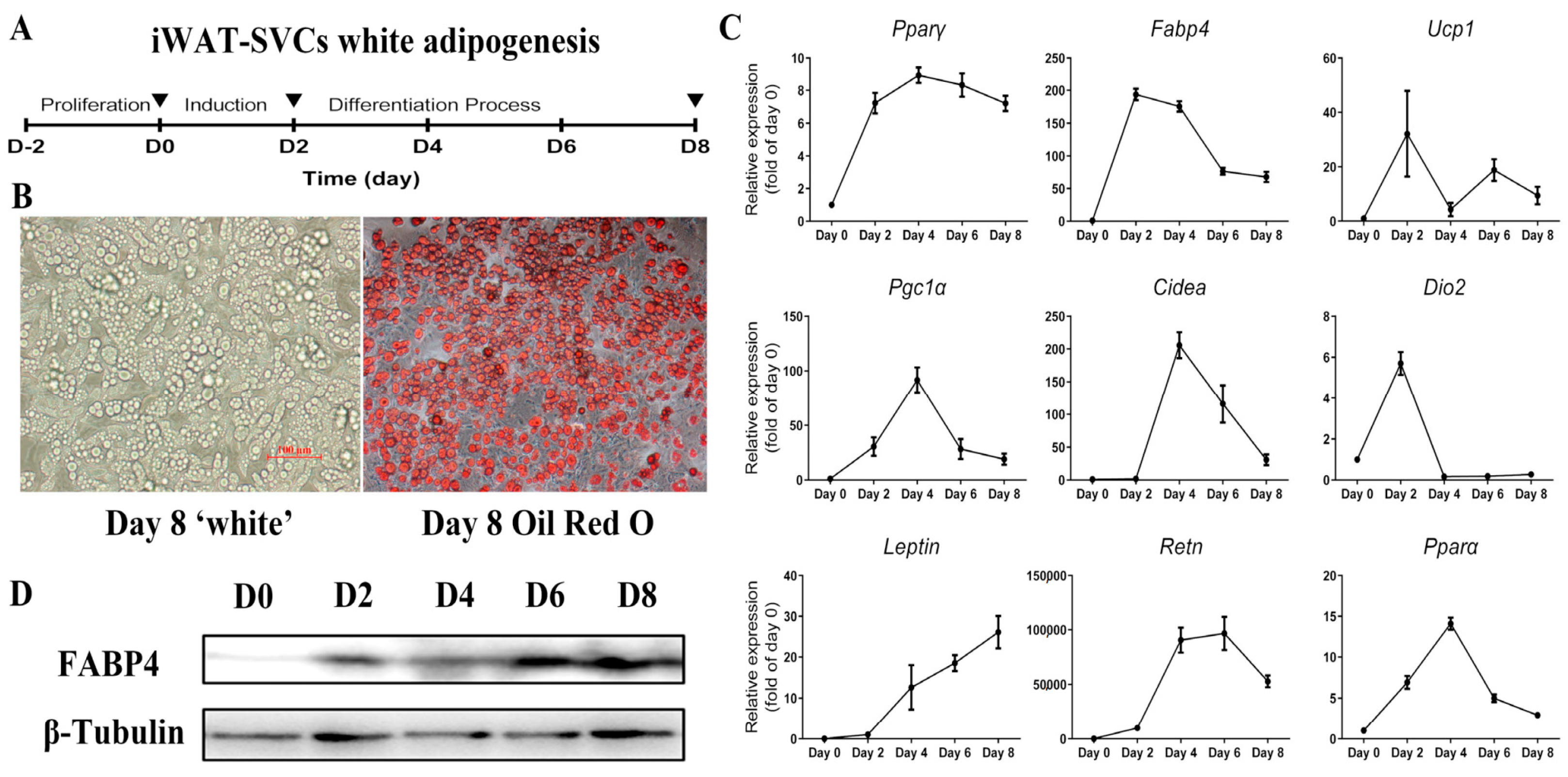
2.1. Lacked Thermogenic Characteristics Under Standard White Adipogenic Conditions
2.2. CL316,243 Induced the Cultured Mature White Adipocyte Beiging
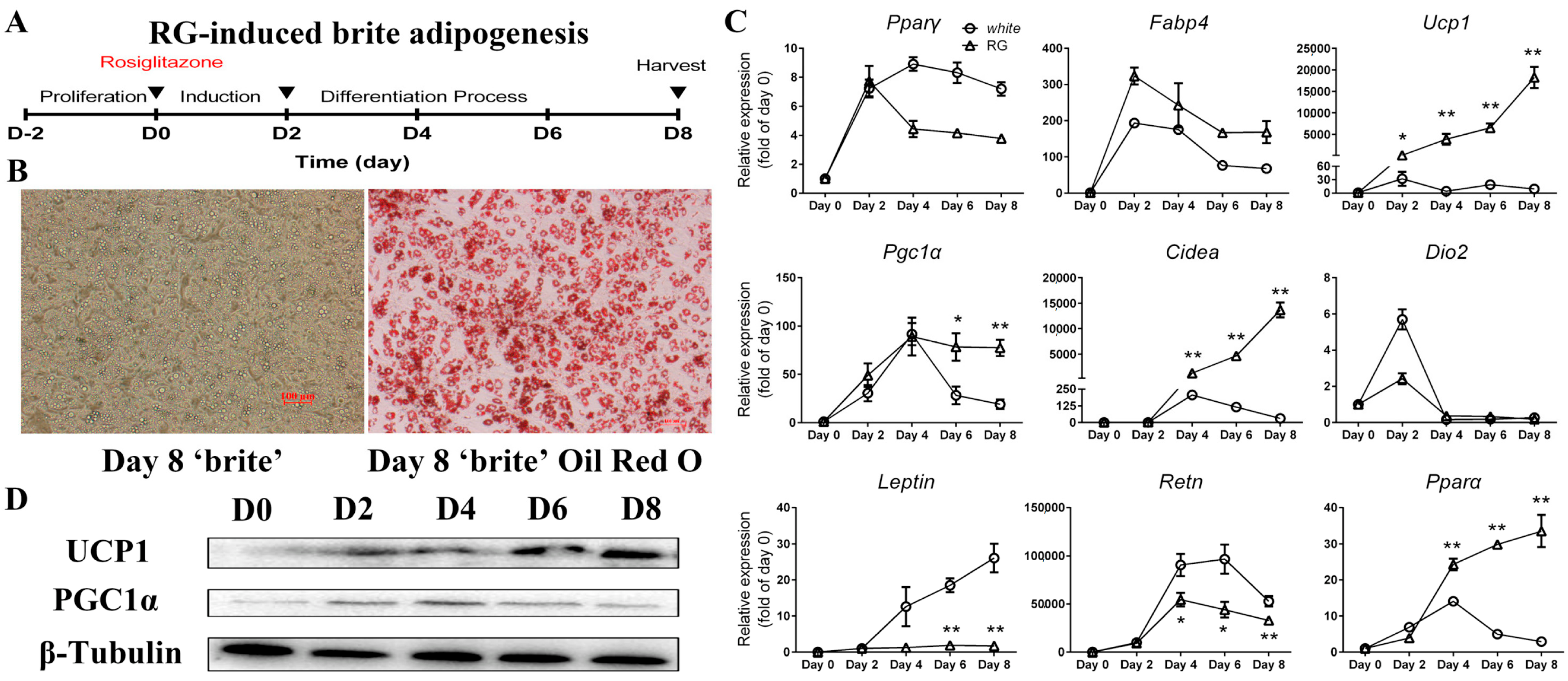
2.3. Differentiation of Mice iWAT-SVCs to Brite Cells in the Presence of Rosiglitazone
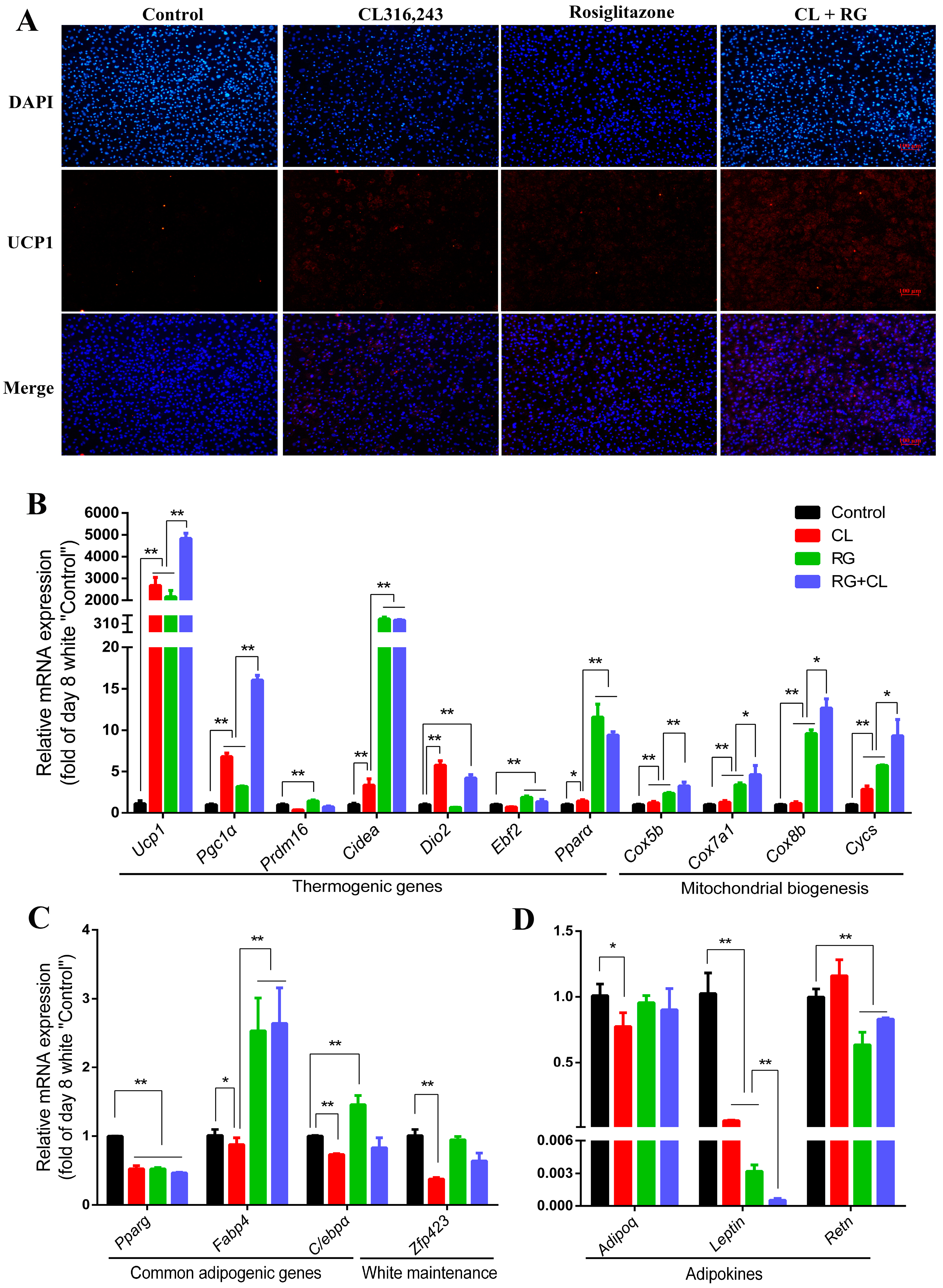
2.4. CL316,243 Further Promoted Thermogenic Gene Expression and Mitochondrial Biogenesis of Rosiglitazone-Induced Brite Adipocytes
3. Discussion
4. Materials and Methods
4.1. Isolation of Adipose SVF and In Vitro Differentiation
4.2. Oil Red O staining
4.3. Gene Expression Analysis
4.4. Protein Analysis
4.5. Immunofluorescence
4.6. Bioinformatic Analysis
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| iWAT-SVCs | Inguinal white adipose tissue derived stromal vascular cells |
| eWAT | Epididymal white adipose tissue |
| Pparα/γ | Peroxisome proliferator activated receptor alpha/gamma |
| Fabp4 | Fatty acid binding protein 4 |
| Ucp1 | Uncoupling protein 1 |
| Pgc1α | Peroxisome proliferative activated receptor, gamma, coactivator 1 alpha |
| Cidea | Cell death-inducing DNA fragmentation factor, alpha subunit-like effector A |
| Dio2 | Deiodinase, iodothyronine, type II |
| Retn | Resistin |
| Prdm16 | PR domain containing 16 |
| Zfp423 | Zinc finger protein 423 |
| Ebf2 | Early B cell factor 2 |
| Cycs | Cytochrome c, somatic |
| Cox5b/7a1/8b | Cytochrome c oxidase subunit 5b/7a1/8b |
| Adrb1 | Adrenergic β1 receptor |
| Tnf (ip3/6) | Tumor necrosis factor (induced protein 3/6) |
| Elovl3 | Elongation of very long chain fatty acids |
| Slc25a20/27a2 | Solute carrier family 25 member 20/ Solute carrier family 27 member 2 |
| Tlr2/7 | Toll-like receptor 2/7 |
| Gpr126 | G protein-coupled receptor 126 |
| Tmem43 | Transmembrane protein 43 |
References
- Deng, T.; Lyon, C.J.; Bergin, S.; Caligiuri, M.A.; Hsueh, W.A. Obesity, inflammation, and cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 421–449. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.S.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and beige fat: Physiological roles beyond heat generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, J.; Seale, P. Beige can be slimming. Science 2010, 328, 1113–1114. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Rong, J.X.; Qiu, Y.; Hansen, M.K.; Zhu, L.; Zhang, V.; Xie, M.; Okamoto, Y.; Mattie, M.D.; Higashiyama, H.; Asano, S.; et al. Adipose mitochondrial biogenesis is suppressed in DB/DB and high-fat diet-fed mice and improved by rosiglitazone. Diabetes 2007, 56, 1751–1760. [Google Scholar] [CrossRef] [PubMed]
- Himms-Hagen, J.; Melnyk, A.; Zingaretti, M.C.; Ceresi, E.; Barbatelli, G.; Cinti, S. Multilocular fat cells in wat of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol. Cell Physiol 2000, 279, 670–681. [Google Scholar]
- Wang, H.; Farmer, S.R. Roscovitine induces browning of white adipose tissue. Diabetes 2015, 64, A548. [Google Scholar]
- Wang, H.; Liu, L.; Lin, J.Z.; Aprahamian, T.R.; Farmer, S.R. Browning of white adipose tissue with roscovitine induces a distinct population of UCP1+ adipocytes. Cell Metab. 2016, 24, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Ying, Z.K.; Cai, M.; Xu, Z.B.; Li, Y.J.; Jiang, S.Y.; Tzan, K.; Wang, A.X.; Parthasarathy, S.; He, G.L.; et al. Exercise ameliorates high-fat diet-induced metabolic and vascular dysfunction, and increases adipocyte progenitor cell population in brown adipose tissue. Am. J. Physiol. Reg. I. 2011, 300, R1115–R1125. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. PRDM16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Bostrom, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostrom, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Aune, U.L.; Ruiz, L.; Kajimura, S. Isolation and differentiation of stromal vascular cells to beige/brite cells. Jove J. Vis. Exp. 2013, 73, e50191. [Google Scholar]
- Luo, X.; Jia, R.; Zhang, Q.L.; Sun, B.; Yan, J.Q. Cold-induced browning dynamically alters the expression profiles of inflammatory adipokines with tissue specificity in mice. Int. J. Mol. Sci. 2016, 17, 795. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Bostrom, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Ohno, H.; Shinoda, K.; Spiegelman, B.M.; Kajimura, S. Ppar gamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012, 15, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Long, J.Z.; Svensson, K.J.; Tsai, L.; Zeng, X.; Roh, H.C.; Kong, X.X.; Rao, R.R.; Lou, J.; Lokurkar, I.; Baur, W.; et al. A smooth muscle-like origin for beige adipocytes. Cell Metab. 2014, 19, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Nogusa, Y.; Shinoda, K.; Suzuki, K.; Bannai, M.; Kajimura, S. A synergistic antiobesity effect by a combination of capsinoids and cold temperature through promoting beige adipocyte biogenesis. Diabetes 2016, 65, 1410–1423. [Google Scholar] [CrossRef] [PubMed]
- Uldry, M.; Yang, W.L.; St-Pierre, J.; Lin, J.D.; Seale, P.; Spiegelman, B.M. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006, 3, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Liu, Q.; Salter, A.M.; Lomax, M.A. Synergism between CAMP and PPARγ signalling in the initiation of UCP1 gene expression in HIB1B brown adipocytes. Ppar. Res. 2013, 2013, 476049. [Google Scholar] [CrossRef] [PubMed]
- Petrovic, N.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Thermogenically competent nonadrenergic recruitment in brown preadipocytes by a PPARγ agonist. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E287–E296. [Google Scholar]
- Kissig, M.; Shapira, S.N.; Seale, P. Snapshot: Brown and beige adipose thermogenesis. Cell 2016, 166, 258. [Google Scholar] [CrossRef] [PubMed]
- Steppan, C.M.; Bailey, S.T.; Bhat, S.; Brown, E.J.; Banerjee, R.R.; Wright, C.M.; Patel, H.R.; Ahima, R.S.; Lazar, M.A. The hormone resistin links obesity to diabetes. Nature 2001, 409, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Kamon, J.; Waki, H.; Terauchi, Y.; Kubota, N.; Hara, K.; Mori, Y.; Ide, T.; Murakami, K.; Tsuboyama-Kasaoka, N.; et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001, 7, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Silha, J.V.; Krsek, M.; Skrha, J.V.; Sucharda, P.; Nyomba, B.L.G.; Murphy, L.J. Plasma resistin, adiponectin and leptin levels in lean and obese subjects: Correlations with insulin resistance. Eur. J. Endocrinol. 2003, 149, 331–335. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-L.; Li, X.; Jiang, T.-T.; Fan, J.-M.; Zheng, X.-L.; Shi, X.-E.; Yu, T.-Y.; Chu, G.-Y.; Yang, G.-S. An Additive Effect of Promoting Thermogenic Gene Expression in Mice Adipose-Derived Stromal Vascular Cells by Combination of Rosiglitazone and CL316,243. Int. J. Mol. Sci. 2017, 18, 1002. https://doi.org/10.3390/ijms18051002
Li Y-L, Li X, Jiang T-T, Fan J-M, Zheng X-L, Shi X-E, Yu T-Y, Chu G-Y, Yang G-S. An Additive Effect of Promoting Thermogenic Gene Expression in Mice Adipose-Derived Stromal Vascular Cells by Combination of Rosiglitazone and CL316,243. International Journal of Molecular Sciences. 2017; 18(5):1002. https://doi.org/10.3390/ijms18051002
Chicago/Turabian StyleLi, You-Lei, Xiao Li, Tian-Tuan Jiang, Jia-Min Fan, Xue-Li Zheng, Xin-E Shi, Tai-Yong Yu, Gui-Yan Chu, and Gong-She Yang. 2017. "An Additive Effect of Promoting Thermogenic Gene Expression in Mice Adipose-Derived Stromal Vascular Cells by Combination of Rosiglitazone and CL316,243" International Journal of Molecular Sciences 18, no. 5: 1002. https://doi.org/10.3390/ijms18051002