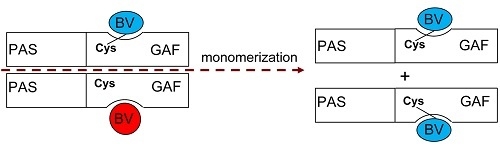
Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome
Abstract
:1. Introduction
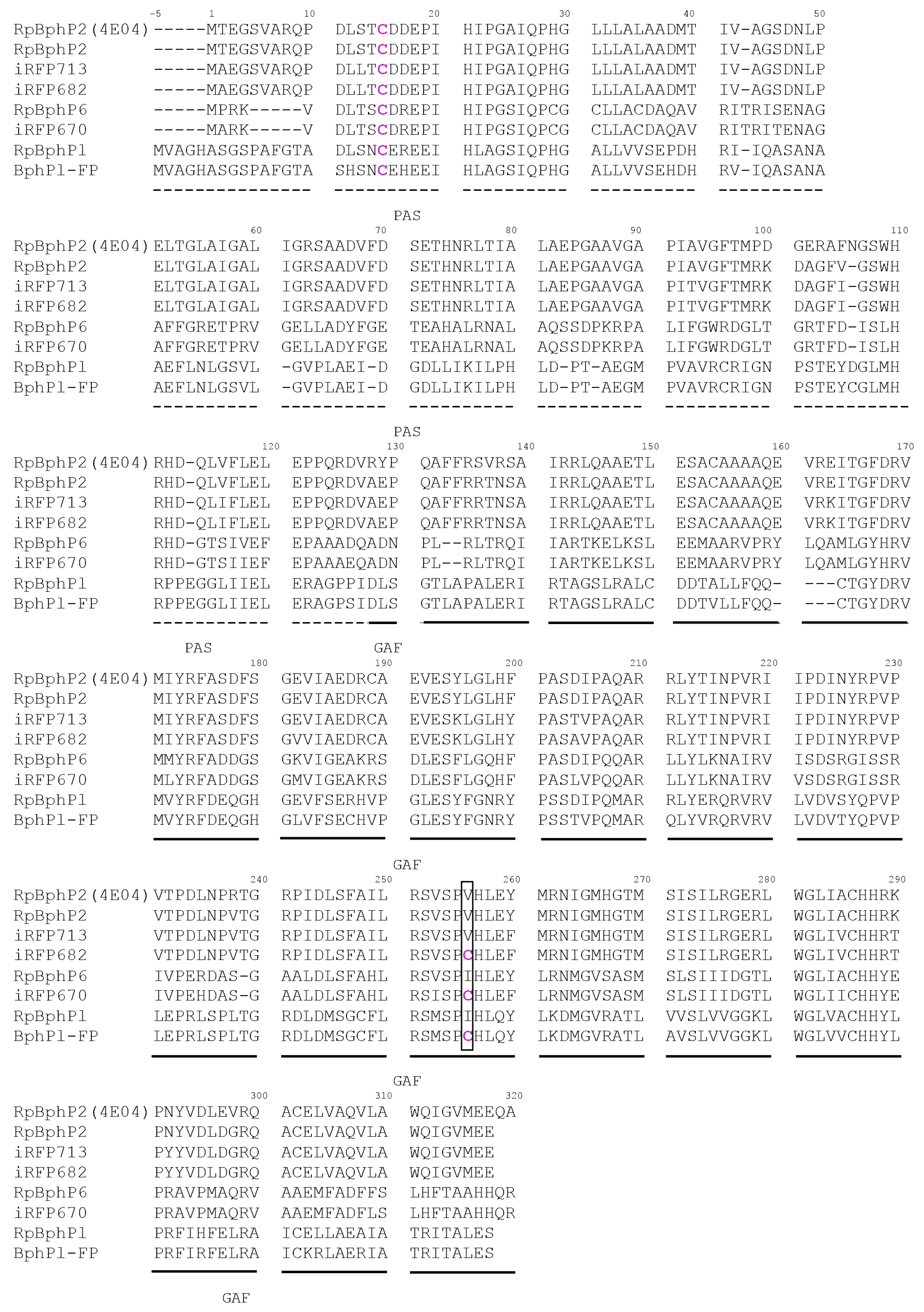
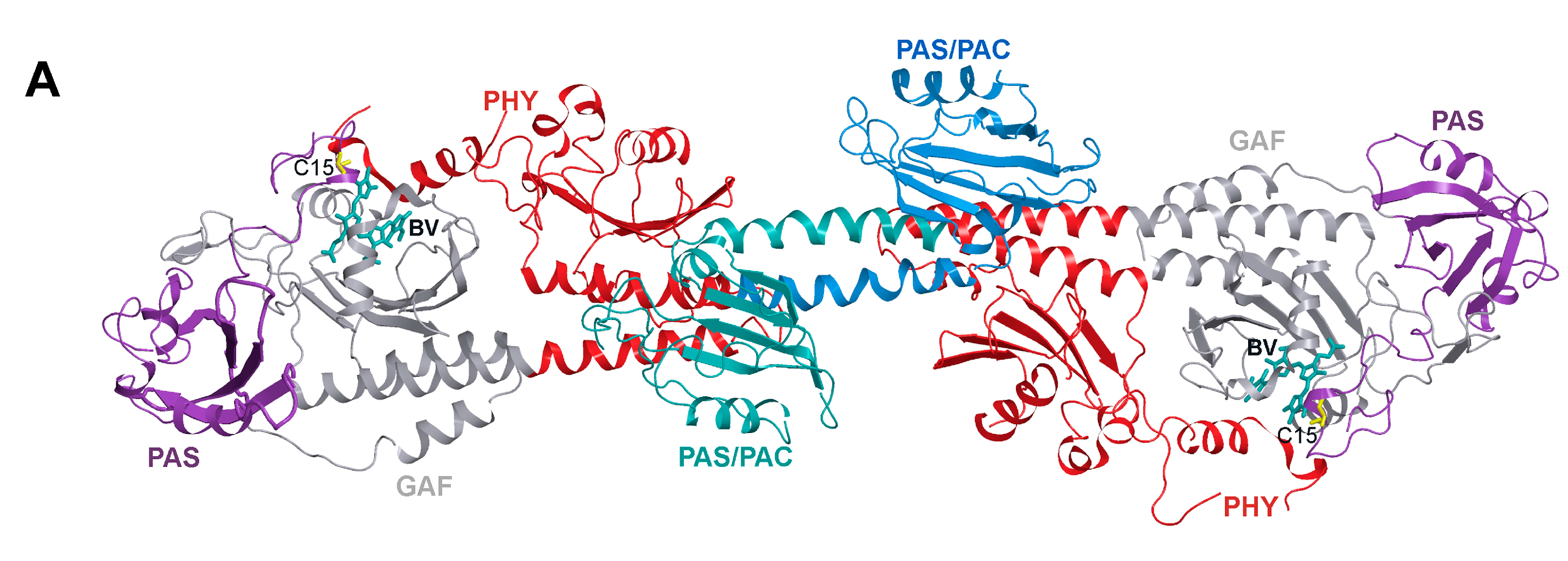
2. Results and Discussion
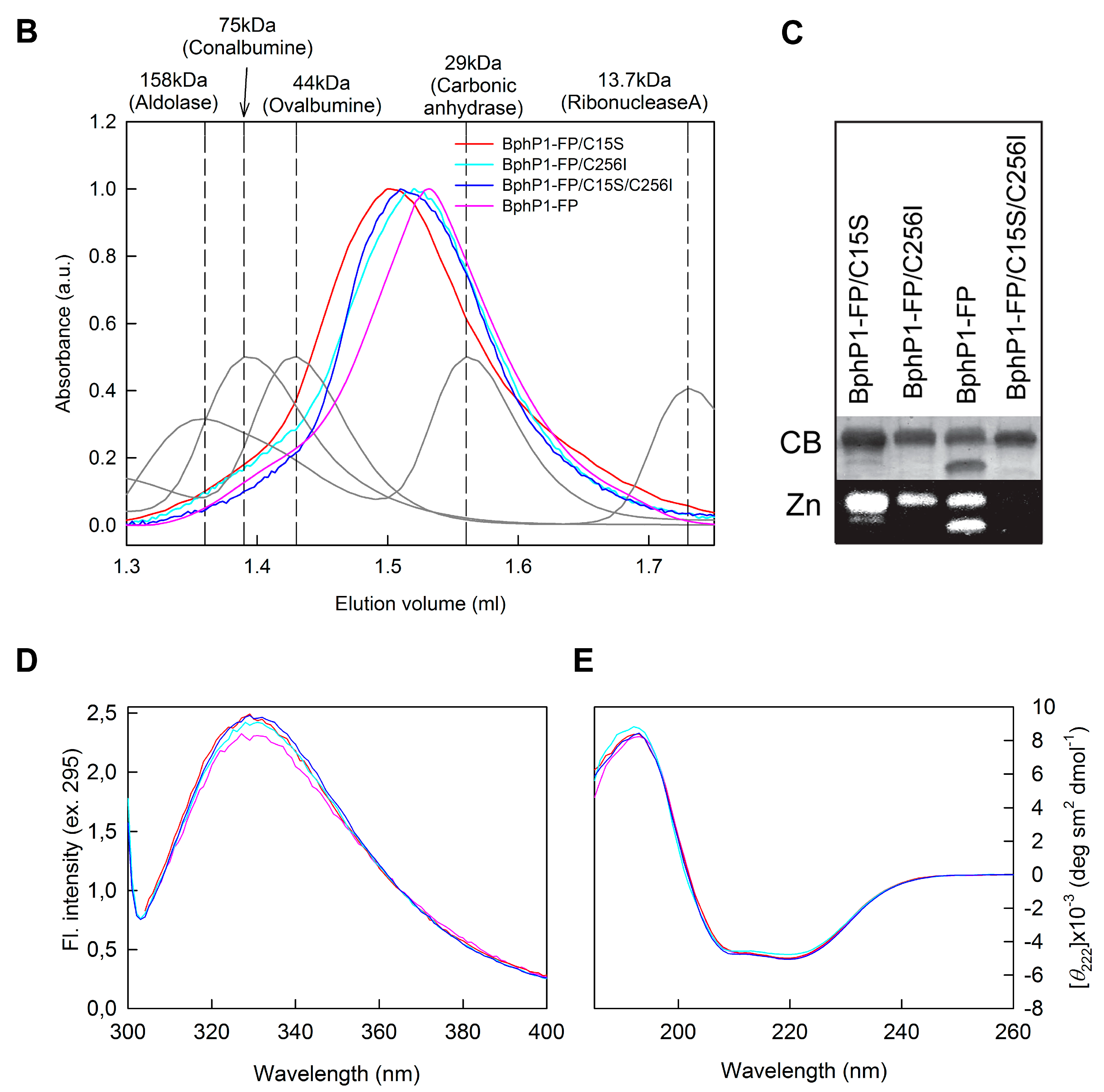
2.1. Properties of Fluorescent Protein BphP1-FP Engineered From Bacterial Phytochrome RpBphP1 and Its Mutants
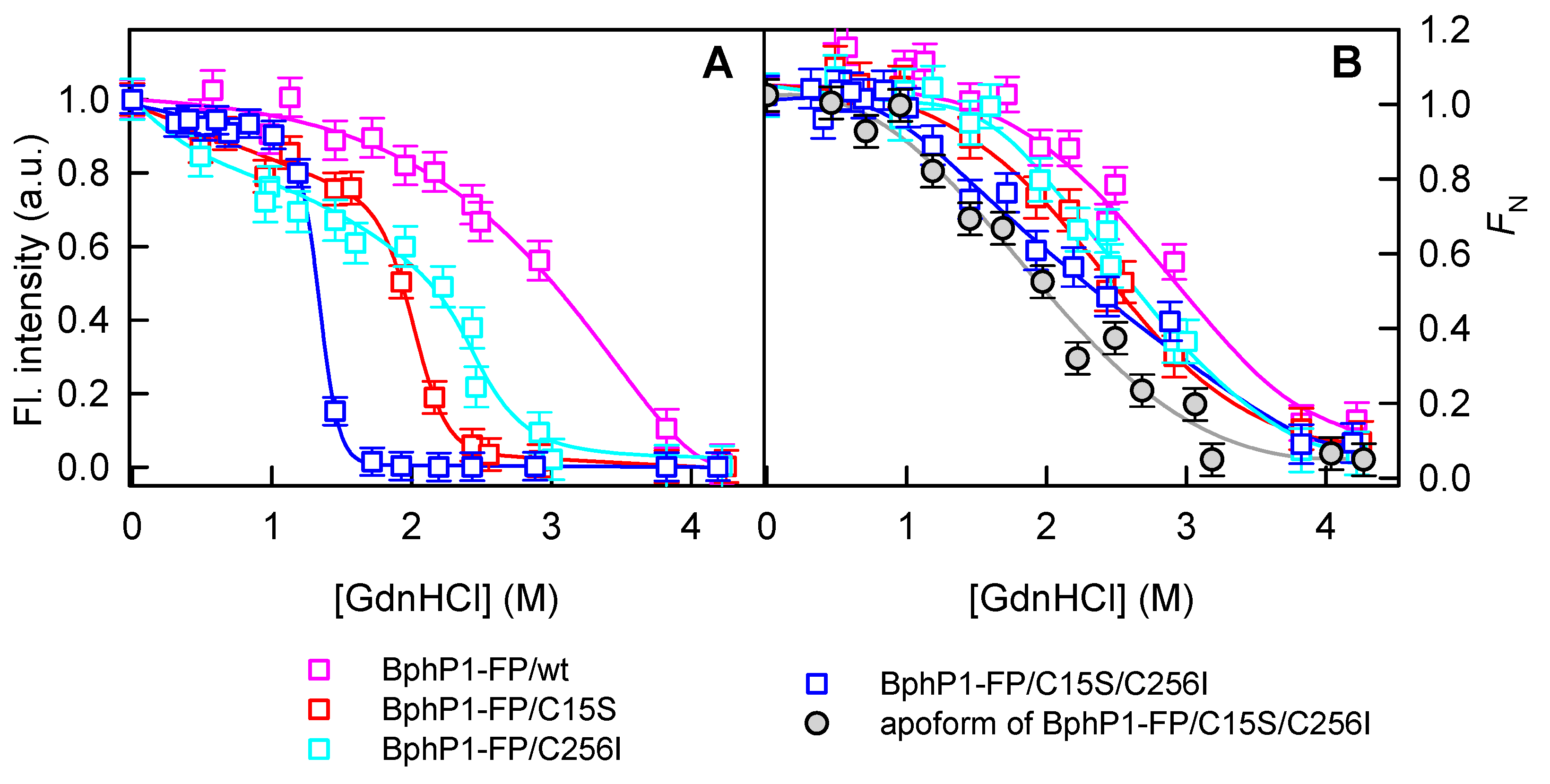
2.2. Properties of BphP1-FP and Its Mutants in Denaturant
3. Materials and Methods
3.1. Plasmids, Mutagenesis, Protein Expression and Purification
3.2. Spectral and Biochemical Characterization of Purified Proteins
3.3. Protein Unfolding Assay
3.4. Circular Dichroism Measurements
3.5. Fitting of Denaturation Curves
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Piatkevich, K.D.; Subach, F.V.; Verkhusha, V.V. Engineering of bacterial phytochromes for near-infrared imaging, sensing, and light-control in mammals. Chem. Soc. Rev. 2013, 42, 3441–3452. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Verkhusha, V.V. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr. Opin. Chem. Biol. 2015, 27, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Li, W.; Zhang, Z.; Liu, S.; Zhang, X.; Zhang, X.E.; Cui, Z. Novel near-infrared BiFC systems from a bacterial phytochrome for imaging protein interactions and drug evaluation under physiological conditions. Biomaterials 2015, 48, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Verkhusha, V.V. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem. Biol. 2013, 20, 1078–1086. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Emelyanov, A.V.; Brenowitz, M.; Guo, P.; Verkhusha, V.V. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 2016, 7, 12405. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, D.M.; Grossman, A.R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996, 273, 1409–1412. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Swem, L.R.; Rushing, B.G.; Devanathan, S.; Tollin, G.; Bauer, C.E. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 1999, 285, 406–409. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.J.; Vener, A.V.; Vierstra, R.D. Bacteriophytochromes: Phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 1999, 286, 2517–2520. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, S.; Suzuki, F.; Fujita, H.; Geng, X.X.; Ikeuchi, M. Novel putative photoreceptor and regulatory genes required for the positive phototactic movement of the unicellular motile cyanobacterium synechocystis sp. PCC 6803. Plant. Cell Physiol. 2000, 41, 1299–1304. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Fardoux, J.; Fourrier, N.; Hannibal, L.; Genty, B.; Bouyer, P.; Dreyfus, B.; Vermeglio, A. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 2002, 417, 202–205. [Google Scholar] [CrossRef] [PubMed]
- Bhoo, S.H.; Davis, S.J.; Walker, J.; Karniol, B.; Vierstra, R.D. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 2001, 414, 776–779. [Google Scholar] [CrossRef] [PubMed]
- Giraud, E.; Vermeglio, A. Bacteriophytochromes in anoxygenic photosynthetic bacteria. Photosynth. Res. 2008, 97, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Karniol, B.; Vierstra, R.D. The pair of bacteriophytochromes from agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. USA 2003, 100, 2807–2812. [Google Scholar] [CrossRef] [PubMed]
- Tasler, R.; Moises, T.; Frankenberg-Dinkel, N. Biochemical and spectroscopic characterization of the bacterial phytochrome of pseudomonas aeruginosa. FEBS J. 2005, 272, 1927–1936. [Google Scholar] [CrossRef] [PubMed]
- Rottwinkel, G.; Oberpichler, I.; Lamparter, T. Bathy phytochromes in rhizobial soil bacteria. J. Bacteriol. 2010, 192, 5124–5133. [Google Scholar] [CrossRef] [PubMed]
- Lamparter, T.; Carrascal, M.; Michael, N.; Martinez, E.; Rottwinkel, G.; Abian, J. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N-terminus of agrobacterium phytochrome Agp1. Biochemistry 2004, 43, 3659–3669. [Google Scholar] [CrossRef] [PubMed]
- Lamparter, T.; Michael, N.; Caspani, O.; Miyata, T.; Shirai, K.; Inomata, K. Biliverdin binds covalently to agrobacterium phytochrome Agp1 via its ring A vinyl side chain. J. Biol. Chem. 2003, 278, 33786–33792. [Google Scholar] [CrossRef] [PubMed]
- Kyndt, J.A.; Fitch, J.C.; Meyer, T.E.; Cusanovich, M.A. Thermochromatium tepidum photoactive yellow protein/bacteriophytochrome/diguanylate cyclase: Characterization of the PYP domain. Biochemistry 2005, 44, 4755–4764. [Google Scholar] [CrossRef] [PubMed]
- Tarutina, M.; Ryjenkov, D.A.; Gomelsky, M. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 2006, 281, 34751–34758. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Verkhusha, V.V. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods 2013, 10, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Gustafson, W.C.; Han, C.; Lafaye, C.; Noirclerc-Savoye, M.; Ge, W.P.; Thayer, D.A.; Huang, H.; Kornberg, T.B.; Royant, A.; et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat. Commun. 2014, 5, 3626. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Auldridge, M.E.; Lehtivuori, H.; Ihalainen, J.A.; Forest, K.T. Origins of fluorescence in evolved bacteriophytochromes. J. Biol. Chem. 2014, 289, 32144–32152. [Google Scholar] [CrossRef] [PubMed]
- Auldridge, M.E.; Satyshur, K.A.; Anstrom, D.M.; Forest, K.T. Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 2012, 287, 7000–7009. [Google Scholar] [CrossRef] [PubMed]
- Filonov, G.S.; Piatkevich, K.D.; Ting, L.M.; Zhang, J.; Kim, K.; Verkhusha, V.V. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat. Biotechnol. 2011, 29, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Subach, F.V.; Verkhusha, V.V. Far-red light photoactivatable near-infrared fluorescent proteins engineered from a bacterial phytochrome. Nat. Commun. 2013, 4, 2153. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Royant, A.; Lin, M.Z.; Aguilera, T.A.; Lev-Ram, V.; Steinbach, P.A.; Tsien, R.Y. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 2009, 324, 804–807. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakova, D.M.; Baloban, M.; Pletnev, S.; Malashkevich, V.N.; Xiao, H.; Dauter, Z.; Verkhusha, V.V. Molecular basis of spectral diversity in near-infrared phytochrome-based fluorescent proteins. Chem. Biol. 2015, 22, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Baloban, M.; Bublikov, G.S.; Shcherbakova, D.M.; Stepanenko, O.V.; Turoverov, K.K.; Kuznetsova, I.M.; Verkhusha, V.V. Allosteric effects of chromophore interaction with dimeric near-infrared fluorescent proteins engineered from bacterial phytochromes. Sci. Rep. 2016, 6, 18750. [Google Scholar] [CrossRef] [PubMed]
- Hontani, Y.; Shcherbakova, D.M.; Baloban, M.; Zhu, J.; Verkhusha, V.V.; Kennis, J.T. Bright blue-shifted fluorescent proteins with Cys in the GAF domain engineered from bacterial phytochromes: Fluorescence mechanisms and excited-state dynamics. Sci. Rep. 2016, 6, 37362. [Google Scholar] [CrossRef] [PubMed]
- Bellini, D.; Papiz, M.Z. Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 2012, 20, 1436–1446. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids. Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hsin, J.; Arkhipov, A.; Yin, Y.; Stone, J.E.; Schulten, K. Using VMD: An introductory tutorial. Curr. Protoc. Bioinform. 2008. [Google Scholar] [CrossRef]
- Merritt, E.A.; Bacon, D.J. Raster3D: Photorealistic molecular graphics. Methods Enzymol. 1997, 277, 505–524. [Google Scholar] [PubMed]
- Baloban, M.; Shcherbakova, D.M.; Pletnev, S.; Pletnev, V.Z.; Lagarias, J.C.; Verkhusha, V.V. Designing brighter near-infrared fluorescent proteins: Insights from structural and biochemical studies. Chem. Sci. 2017. [Google Scholar] [CrossRef]
- Wagner, J.R.; Zhang, J.; von Stetten, D.; Gunther, M.; Murgida, D.H.; Mroginski, M.A.; Walker, J.M.; Forest, K.T.; Hildebrandt, P.; Vierstra, R.D. Mutational analysis of deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 2008, 283, 12212–12226. [Google Scholar] [CrossRef] [PubMed]
- Burgie, E.S.; Zhang, J.; Vierstra, R.D. Crystal structure of deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 2016, 24, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Bjorling, A.; Berntsson, O.; Lehtivuori, H.; Niebling, S.; Hoernke, M.; Kosheleva, I.; Henning, R.; Menzel, A.; Ihalainen, J.A.; et al. Signal amplification and transduction in phytochrome photosensors. Nature 2014, 509, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Niebling, S.; Berntsson, O.; Bjorling, A.; Lehtivuori, H.; Hakkanen, H.; Panman, M.; Gustavsson, E.; Hoernke, M.; Newby, G.; et al. Light-induced structural changes in a monomeric bacteriophytochrome. Struct. Dyn. 2016, 3, 054701. [Google Scholar] [CrossRef] [PubMed]
- Takala, H.; Bjorling, A.; Linna, M.; Westenhoff, S.; Ihalainen, J.A. Light-induced changes in the dimerization interface of bacteriophytochromes. J. Biol. Chem. 2015, 290, 16383–16392. [Google Scholar] [CrossRef] [PubMed]
- Stepanenko, O.V.; Stepanenko, O.V.; Bublikov, G.S.; Kuznetsova, I.M.; Verkhusha, V.V.; Turoverov, K.K. Stabilization of structure in near-infrared fluorescent proteins by binding of biliverdin chromophore. J. Mol. Struct. 2016, 1140, 22–31. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Berkelman, T.R.; Lagarias, J.C. Visualization of bilin-linked peptides and proteins in polyacrylamide gels. Anal. Biochem. 1986, 156, 194–201. [Google Scholar] [CrossRef]
- Kuznetsova, I.M.; Sulatskaya, A.I.; Povarova, O.I.; Turoverov, K.K. Reevaluation of ans binding to human and bovine serum albumins: Key role of equilibrium microdialysis in ligand—Receptor binding characterization. PLoS ONE 2012, 7, e40845. [Google Scholar] [CrossRef] [PubMed]
- Fonin, A.V.; Sulatskaya, A.I.; Kuznetsova, I.M.; Turoverov, K.K. Fluorescence of dyes in solutions with high absorbance. Inner filter effect correction. PLoS ONE 2014, 9, e103878. [Google Scholar] [CrossRef] [PubMed]
- Nolting, B. Protein Folding Kinetics. In Biophysical Methods; Springer: Berlin/Heidelberg, Germany, 1999; p. 191. [Google Scholar]
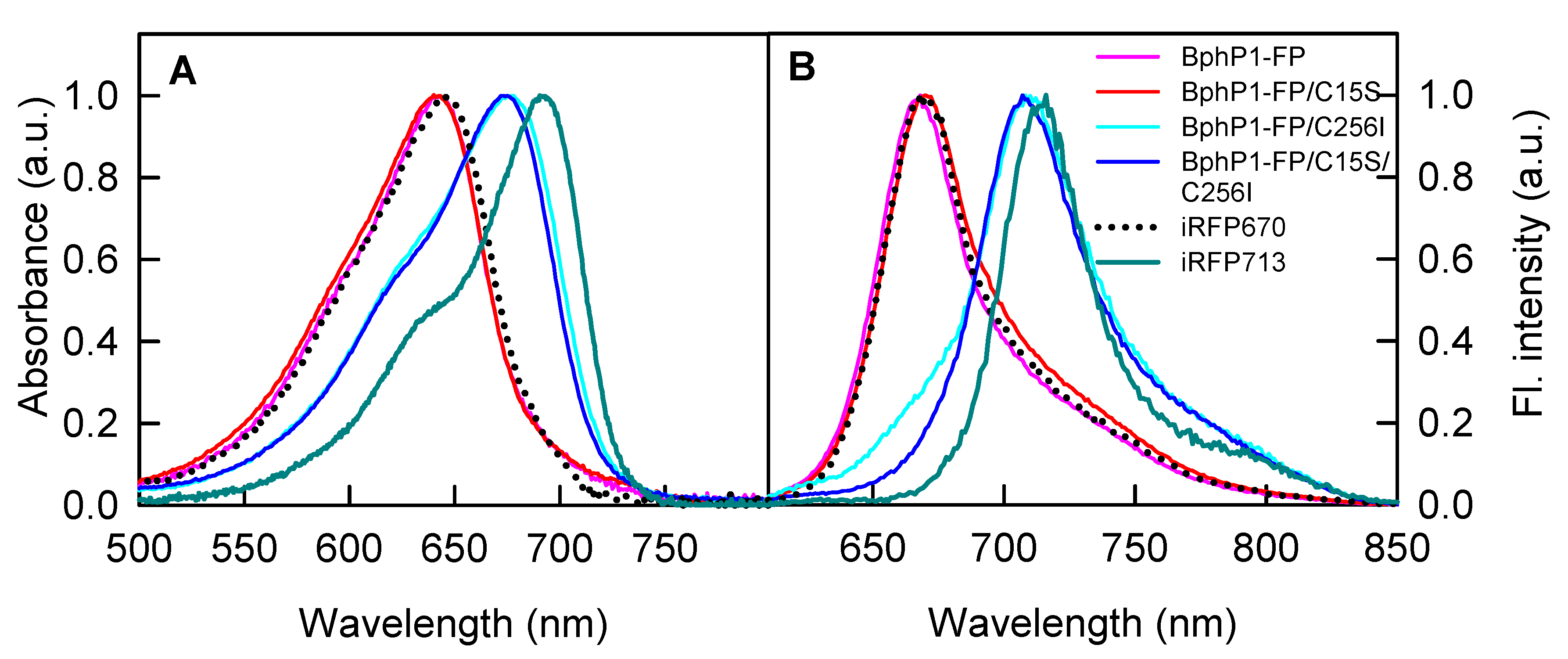


| NIR FP | Absorbance Maximum (nm) | FWHM 1 of Absorption Spectrum (nm) | Emission Maximum (nm) | FWHM of Emission Spectrum (nm) | Extinction Coefficient (M−1·cm−1) | Quantum Yield (%) | Molecular Brightness 2 vs. iRFP713 (%) |
|---|---|---|---|---|---|---|---|
| iRFP713 3 | 692 | 66 | 713 | 38 | 98,000 | 6.3 | 100 |
| iRFP670 3 | 644 | 78 | 670 | 42 | 110,000 | 12.2 | 217 |
| BphP1-FP | 643 | 77 | 668 | 42 | 66,800 | 13.8 | 149 |
| BphP1-FP/C15S | 643 | 80 | 670 | 46 | 71,100 | 12.1 | 139 |
| BphP1-FP/C256I | 677 | 89 | 708 | 54 | 63,400 | 3.8 | 39 |
| BphP1-FP/C15S/C256I | 675 | 85 | 707 | 50 | 69,200 | 4.6 | 52 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stepanenko, O.V.; Stepanenko, O.V.; Kuznetsova, I.M.; Shcherbakova, D.M.; Verkhusha, V.V.; Turoverov, K.K. Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome. Int. J. Mol. Sci. 2017, 18, 1009. https://doi.org/10.3390/ijms18051009
Stepanenko OV, Stepanenko OV, Kuznetsova IM, Shcherbakova DM, Verkhusha VV, Turoverov KK. Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome. International Journal of Molecular Sciences. 2017; 18(5):1009. https://doi.org/10.3390/ijms18051009
Chicago/Turabian StyleStepanenko, Olesya V., Olga V. Stepanenko, Irina M. Kuznetsova, Daria M. Shcherbakova, Vladislav V. Verkhusha, and Konstantin K. Turoverov. 2017. "Interaction of Biliverdin Chromophore with Near-Infrared Fluorescent Protein BphP1-FP Engineered from Bacterial Phytochrome" International Journal of Molecular Sciences 18, no. 5: 1009. https://doi.org/10.3390/ijms18051009