Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy
Abstract
:1. Introduction
2. Results
2.1. Study Population
2.2. Role of Clinical Parameters and Circulating Biomarkers in Prognosis
2.3. Circulating Biomarkers and Treatment
3. Discussion
4. Materials and Methods
4.1. Patients’ Enrollment
4.2. Circulating Free DNA Isolation and Quantification
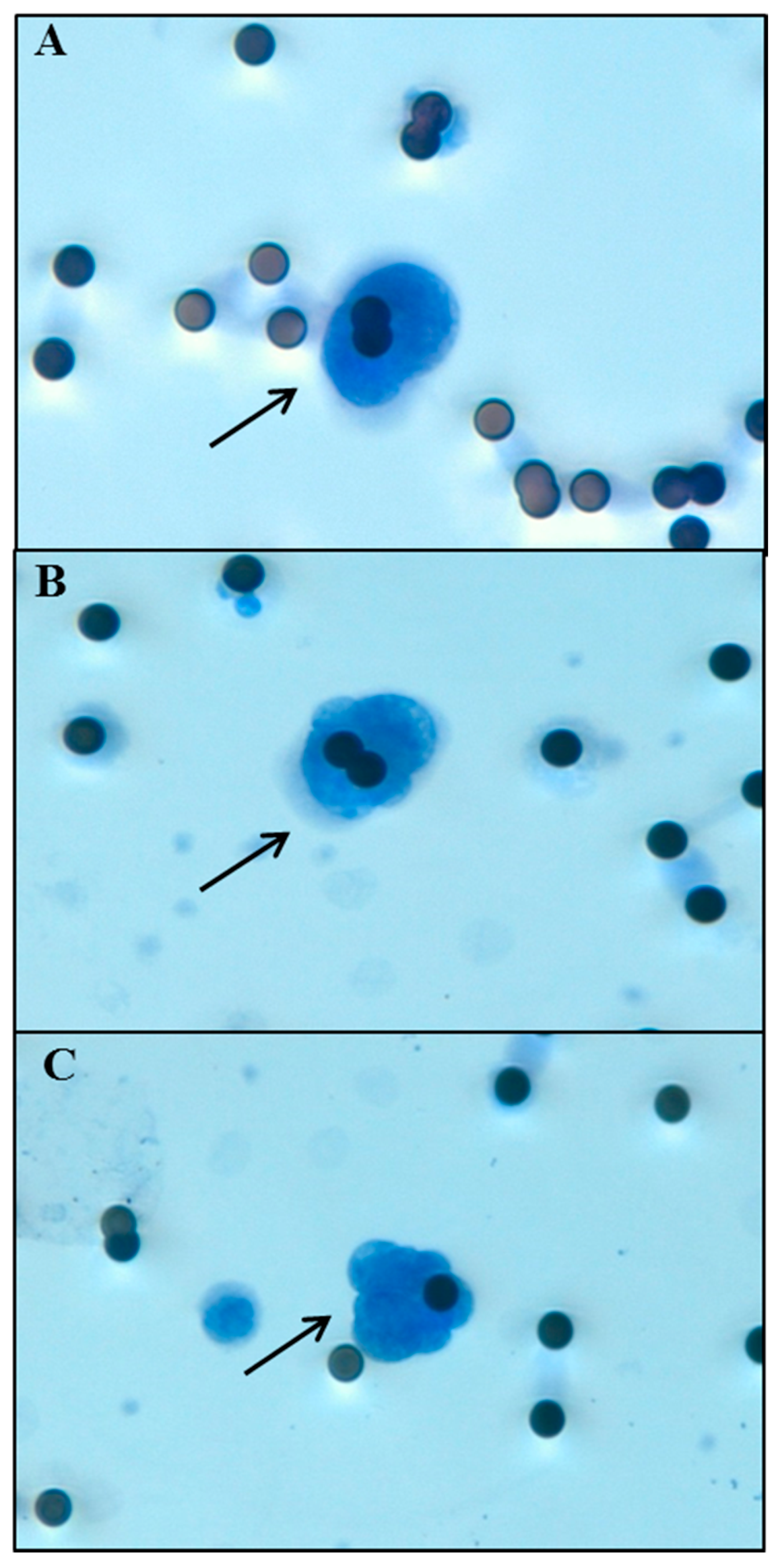
4.3. Circulating Tumor Cell Isolation
4.4. Statistical Methods
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ALK | Anaplastic lymphoma kinase |
| BOR | Best Overall Response |
| cfDNA | cell-free tumor DNA |
| CL | Confidence Limits |
| CT | Cycle Threshold |
| CTCs | Circulating Tumor Cells |
| CT-scan | Computed Tomography |
| ECOG PS | Eastern Cooperative Oncology Group Performance Status |
| EDTA | Ethylenediamine tetraacetic acid |
| EGFR | Epidermal growth factor receptor |
| EpCAM | Epithelial Cell Adhesion Molecule |
| GM | Geometrical Mean |
| H&E | Haematoxylin-Eosin |
| HR | Hazard Ratio |
| hTERT | human Telomerase Reverse Transcriptase |
| NSCLC | Non-small cell lung cancer |
| OS | Overall Survival |
| PD | Progressive Disease |
| PFS | Progression-Free Survival |
| PR | Partial Response |
| qPCR | quantitative PCR |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| SD | Stable Disease |
References
- National Cancer Institute. Non-Small Cell Lung Cancer Treatment (PDQ®). National Cancer Institute Website. Available online: https://www.cancer.gov/types/lung/hp/non-small-cell-lung-treatment-pdq (accessed on 20 January 2017).
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Ries, L.A.G.; Eisner, M.P.; Kosary, C.L.; Hankey, B.F.; Miller, B.A.; Clegg, L.; Edwards, B.K. Cancer Statistics Review 1975–2002; National Cancer Institute: Bethesda, MD, USA, 2005. [Google Scholar]
- Goldstraw, P.; Crowley, J.; Chansky, K.; Giroux, D.J.; Groome, P.A.; Rami-Porta, R.; Postmus, P.E.; Rusch, V.; Sobin, L.; International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions. The IASLC Lung Cancer Staging Project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J. Thorac. Oncol. 2007, 2, 706–714. [Google Scholar] [PubMed]
- Rodrigues-Pereira, J.; Kim, J.H.; Magallanes, M.; Lee, D.H.; Wang, J.; Ganju, V.; Martínez-Barrera, L.; Barraclough, H.; van Kooten, M.; Orlando, M. A randomized phase 3 trial comparing pemetrexed/carboplatin and docetaxel/carboplatin as first-line treatment for advanced, nonsquamous non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Moro-Sibilot, D.; Smit, E.; de Castro Carpeño, J.; Lesniewski-Kmak, K.; Aerts, J.; Villatoro, R.; Kraaij, K.; Nacerddine, K.; Dyachkova, Y.; Smith, K.T.; et al. Outcomes and resource use of non-small cell lung cancer (NSCLC) patients treated with first-line platinum-based chemotherapy across Europe: FRAME prospective observational study. Lung Cancer 2015, 88, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xia, W.; Lv, Z.; Xin, Y.; Ni, C.; Yang, L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell. Physiol. Biochem. 2017, 41, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Ishikawa, Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J. Clin. Med. 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Stroun, M.; Maurice, P.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Lefort, F.; Rossier, A.; Chen, X.Q.; Anker, P. The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 2000, 906, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Bednarz-Knoll, N.; Alix-Panabières, C.; Pantel, K. Plasticity of disseminating cancer cells in patients with epithelial malignancies. Cancer Metastasis Rev. 2012, 31, 673–687. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- De Bono, J.S.; Scher, H.I.; Montgomery, R.B.; Parker, C.; Miller, M.C.; Tissing, H.; Doyle, G.V.; Terstappen, L.W.; Pienta, K.J.; Raghavan, D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 2008, 14, 6302–6309. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.; Mitchell, E.; Miller, M.C.; et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. Clin. Oncol. 2008, 26, 3213–3221. [Google Scholar] [CrossRef] [PubMed]
- Desitter, I.; Guerrouahen, B.S.; Benali-Furet, N.; Wechsler, J.; Jänne, P.A.; Kuang, Y.; Yanagita, M.; Wang, L.; Berkowitz, J.A.; Distel, R.J.; et al. A new device for rapid isolation by size and characterization of rare circulating tumor cells. Anticancer Res. 2011, 31, 427–441. [Google Scholar] [PubMed]
- Truini, A.; Alama, A.; Dal Bello, M.G.; Coco, S.; Vanni, I.; Rijavec, E.; Genova, C.; Barletta, G.; Biello, F.; Grossi, F. Clinical applications of circulating tumor cells in lung cancer patients by CellSearch System. Front. Oncol. 2014, 4, 242. [Google Scholar] [CrossRef] [PubMed]
- Alama, A.; Truini, A.; Coco, S.; Genova, C.; Grossi, F. Prognostic and predictive relevance of circulating tumor cells in patients with non-small-cell lung cancer. Drug Discov. Today 2014, 19, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Ai, B.; Liu, H.; Huang, Y.; Peng, P. Circulating cell-free DNA as a prognostic and predictive biomarker in non-small cell lung cancer. Oncotarget 2016, 7, 44583–44595. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.X.; Liu, Z.; Jiang, H.L.; Pan, H.M.; Han, W.D. Circulating tumor cells predict survival benefit from chemotherapy in patients with lung cancer. Oncotarget 2016, 7, 67586–67596. [Google Scholar] [CrossRef] [PubMed]
- Arcila, M.E.; Nafa, K.; Chaft, J.E.; Rekhtman, N.; Lau, C.; Reva, B.A.; Zakowski, M.F.; Kris, M.G.; Ladanyi, M. EGFR exon 20 insertion mutations in lung adenocarcinomas: Prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol. Cancer Ther. 2013, 12, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Vanni, I.; Alama, A.; Grossi, F.; Dal Bello, MG.; Coco, S. Exosomes: A new horizon in lung cancer. Drug Discov. Today 2017. [Google Scholar] [CrossRef] [PubMed]
- Fiammengo, R. Can nanotechnology improve cancer diagnosis through miRNA detection? Biomark. Med. 2017, 11, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Gold, B.; Cankovic, M.; Furtado, L.V.; Meier, F.; Gocke, C.D. Do circulating tumor cells, exosomes, and circulating tumor nucleic acids have clinical utility? A report of the association for molecular pathology. J. Mol. Diagn. 2015, 17, 209–224. [Google Scholar] [CrossRef] [PubMed]
- Tissot, C.; Toffart, A.C.; Villar, S.; Souquet, P.J.; Merle, P.; Moro-Sibilot, D.; Pérol, M.; Zavadil, J.; Brambilla, C.; Olivier, M.; et al. Circulating free DNA concentration is an independent prognostic biomarker in lung cancer. Eur. Respir. J. 2015, 46, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Dowler Nygaard, A.; Spindler, K.L.; Pallisgaard, N.; Andersen, R.F.; Jakobsen, A. Levels of cell-free DNA and plasma KRAS during treatment of advanced NSCLC. Oncol. Rep. 2014, 31, 969–974. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Yoon, K.A.; Han, J.Y.; Kim, H.T.; Yun, T.; Lee, G.K.; Kim, H.Y.; Lee, J.S. Circulating cell-free DNA in plasma of never smokers with advanced lung adenocarcinoma receiving gefitinib or standard chemotherapy as first-line therapy. Clin. Cancer Res. 2011, 17, 5179–5187. [Google Scholar] [CrossRef] [PubMed]
- Vinayanuwattikun, C.; Winayanuwattikun, P.; Chantranuwat, P.; Mutirangura, A.; Sriuranpong, V. The impact of non-tumor-derived circulating nucleic acids implicates the prognosis of non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2013, 139, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Sirera, R.; Bremnes, R.M.; Cabrera, A.; Jantus-Lewintre, E.; Sanmartín, E.; Blasco, A.; Del Pozo, N.; Rosell, R.; Guijarro, R.; Galbis, J.; et al. Circulating DNA is a useful prognostic factor in patients with advanced non-small cell lung cancer. J. Thorac. Oncol. 2011, 6, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Camps, C.; Sirera, R.; Bremnes, R.M.; Ródenas, V.; Blasco, A.; Safont, M.J.; Garde, J.; Juarez, A.; Caballero, C.; Sanchez, J.J.; et al. Quantification in the serum of the catalytic fraction of reverse telomerase: A useful prognostic factor in advanced non-small cell lung cancer. Anticancer Res. 2006, 26, 4905–4909. [Google Scholar] [PubMed]
- Li, B.T.; Drilon, A.; Johnson, M.L.; Hsu, M.; Sima, C.S.; McGinn, C.; Sugita, H.; Kris, M.G.; Azzoli, C.G. A prospective study of total plasma cell-free DNA as a predictive biomarker for response to systemic therapy in patients with advanced non-small-cell lung cancers. Ann. Oncol. 2016, 27, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Hughes, A.; Clack, G.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef] [PubMed]
- Hirose, T.; Murata, Y.; Oki, Y.; Sugiyama, T.; Kusumoto, S.; Ishida, H.; Shirai, T.; Nakashima, M.; Yamaoka, T.; Okuda, K.; et al. Relationship of circulating tumor cells to the effectiveness of cytotoxic chemotherapy in patients with metastatic non-small-cell lung cancer. Oncol. Res. 2012, 20, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Hou, J.M.; Sloane, R.; Lancashire, L.; Priest, L.; Nonaka, D.; Ward, T.H.; Backen, A.; Clack, G.; Hughes, A.; et al. Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J. Thorac. Oncol. 2012, 7, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Muinelo-Romay, L.; Vieito, M.; Abalo, A.; Nocelo, M.A.; Barón, F.; Anido, U.; Brozos, E.; Vázquez, F.; Aguín, S.; Abal, M.; et al. Evaluation of Circulating Tumor Cells and Related Events as Prognostic Factors and Surrogate Biomarkers in Advanced NSCLC Patients Receiving First-Line Systemic Treatment. Cancers 2014, 6, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, Y.; Zhao, J.; Chen, M.; Xu, Y.; Zhong, W.; Xing, J.; Wang, M. Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016, 21, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Dong, F.; Cui, F.; Xu, R.; Tang, X. The role of circulating tumor cells in evaluation of prognosis and treatment response in advanced non-small-cell lung cancer. Cancer Chemother. Pharmacol. 2017, 79, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Juan, O.; Vidal, J.; Gisbert, R.; Muñoz, J.; Maciá, S.; Gómez-Codina, J. Prognostic significance of circulating tumor cells in advanced non-small cell lung cancer patients treated with docetaxel and gemcitabine. Clin. Transl. Oncol. 2014, 16, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Hanssen, A.; Loges, S.; Pantel, K.; Wikman, H. Detection of Circulating Tumor Cells in Non-Small Cell Lung Cancer. Front. Oncol. 2015, 5, 207. [Google Scholar] [CrossRef] [PubMed]
- Messaritakis, I.; Politaki, E.; Plataki, M.; Karavassilis, V.; Kentepozidis, N.; Koinis, F.; Samantas, E.; Georgoulias, V.; Kotsakis, A. Heterogeneity of circulating tumor cells (CTCs) in patients with recurrent small cell lung cancer (SCLC) treated with pazopanib. Lung Cancer 2017, 104, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Bulfoni, M.; Turetta, M.; del Ben, F.; di Loreto, C.; Beltrami, A.P.; Cesselli, D. Dissecting the Heterogeneity of Circulating Tumor Cells in Metastatic Breast Cancer: Going Far Beyond the Needle in the Haystack. Int. J. Mol. Sci. 2016, 17, 1775. [Google Scholar] [CrossRef] [PubMed]
- Mascalchi, M.; Falchini, M.; Maddau, C.; Salvianti, F.; Nistri, M.; Bertelli, E.; Sali, L.; Zuccherelli, S.; Vella, A.; Matucci, M.; et al. Prevalence and number of circulating tumour cells and microemboli at diagnosis of advanced NSCLC. J. Cancer Res. Clin. Oncol. 2016, 142, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Guleria, R.; Singh, V.; Bharti, A.C.; Mohan, A.; Das, B.C. Plasma DNA level in predicting therapeutic efficacy in advanced nonsmall cell lung cancer. Eur. Respir. J. 2010, 36, 885–892. [Google Scholar] [CrossRef] [PubMed]
- Gautschi, O.; Bigosch, C.; Huegli, B.; Jermann, M.; Marx, A.; Chassé, E.; Ratschiller, D.; Weder, W.; Joerger, M.; Betticher, D.C.; et al. Circulating deoxyribonucleic acid as prognostic marker in non-small-cell lung cancer patients undergoing chemotherapy. J. Clin. Oncol. 2004, 22, 4157–4164. [Google Scholar] [CrossRef] [PubMed]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/NCT02055144 (accessed on 20 January 2017).
- Genova, C.; Rijavec, E.; Truini, A.; Coco, S.; Sini, C.; Barletta, G.; Dal Bello, M.G.; Alama, A.; Savarino, G.; Pronzato, P.; et al. Pemetrexed for the treatment of non-small cell lung cancer. Expert Opin. Pharmacother. 2013, 14, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Freidin, M.B.; Tay, A.; Freydina, D.V.; Chudasama, D.; Nicholson, A.G.; Rice, A.; Anikin, V.; Lim, E. An assessment of diagnostic performance of a filter-based antibody-independent peripheral blood circulating tumour cell capture paired with cytomorphologic criteria for the diagnosis of cancer. Lung Cancer 2014, 85, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Marubini, E.; Valsecchi, M.G. Analysing Survival Data from Clinical Trials and Observational Studies. In Statistics in Practice, 1st ed.; John Wiley & Sons: West Sussex, UK, 2004; Volume 15, pp. 1–432. [Google Scholar]
| Characteristics | |
| Number of patients | 73 |
| Age at first cycle (years) | |
| Median | 67 |
| Range | 44–80 |
| Gender n (%) | |
| Male | 50 (68.5%) |
| Female | 23 (31.5%) |
| Histology n (%) | |
| Adenocarcinoma | 59 (80.8%) |
| Squamous cell carcinoma | 14 (19.2%) |
| Metastases n (%) | |
| M1a | 23 (31.5%) |
| M1b | 50 (68.5%) |
| Evidence of Brain Metastases n (%) | |
| Yes | 19 (26%) |
| No | 54 (74%) |
| ECOG PS n (%) | |
| 0 | 21 (28.8%) |
| 1–2 | 52 (71.2%) |
| Smoking status n (%) | |
| Current smoker | 31 (42.5%) |
| Former smoker | 36 (49.3%) |
| Never smoker | 6 (8.2%) |
| Factors & Levels | Number of Patients | Number of Deaths (%) | 18-Month OS | p-Value | |
|---|---|---|---|---|---|
| Prob. | 95% CL | ||||
| Baseline cfDNA | |||||
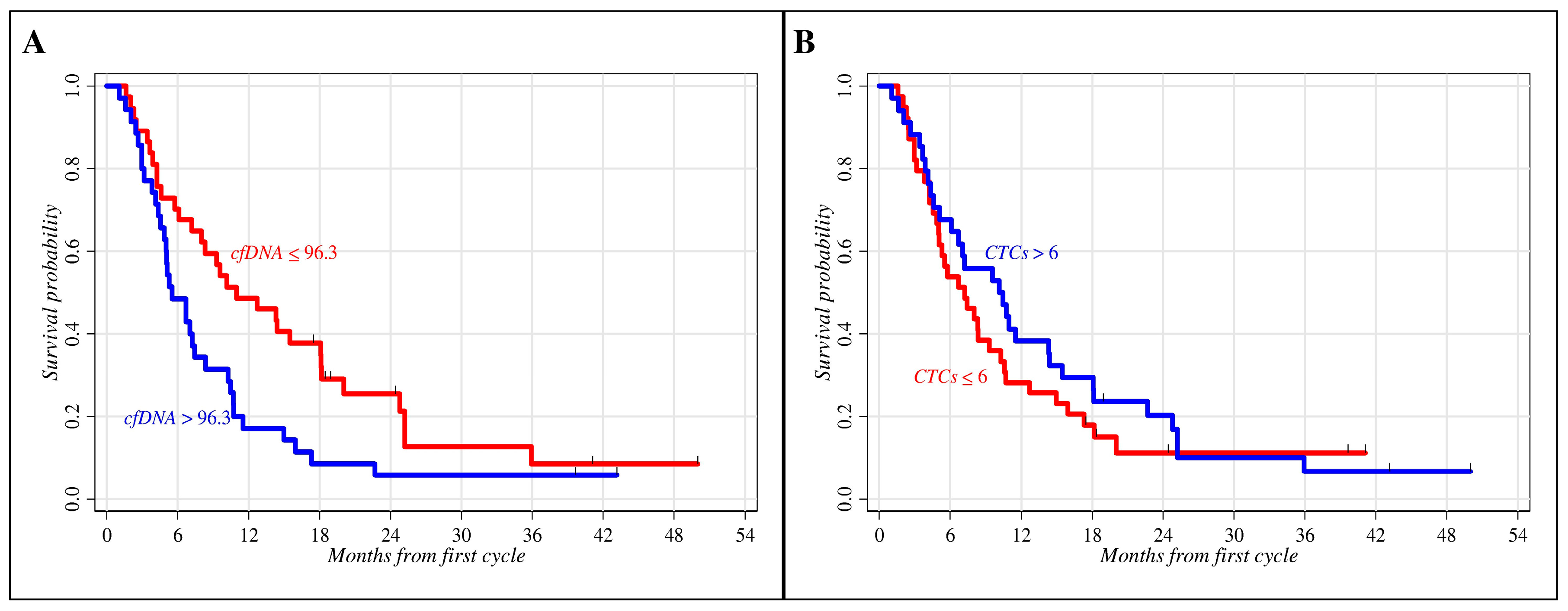
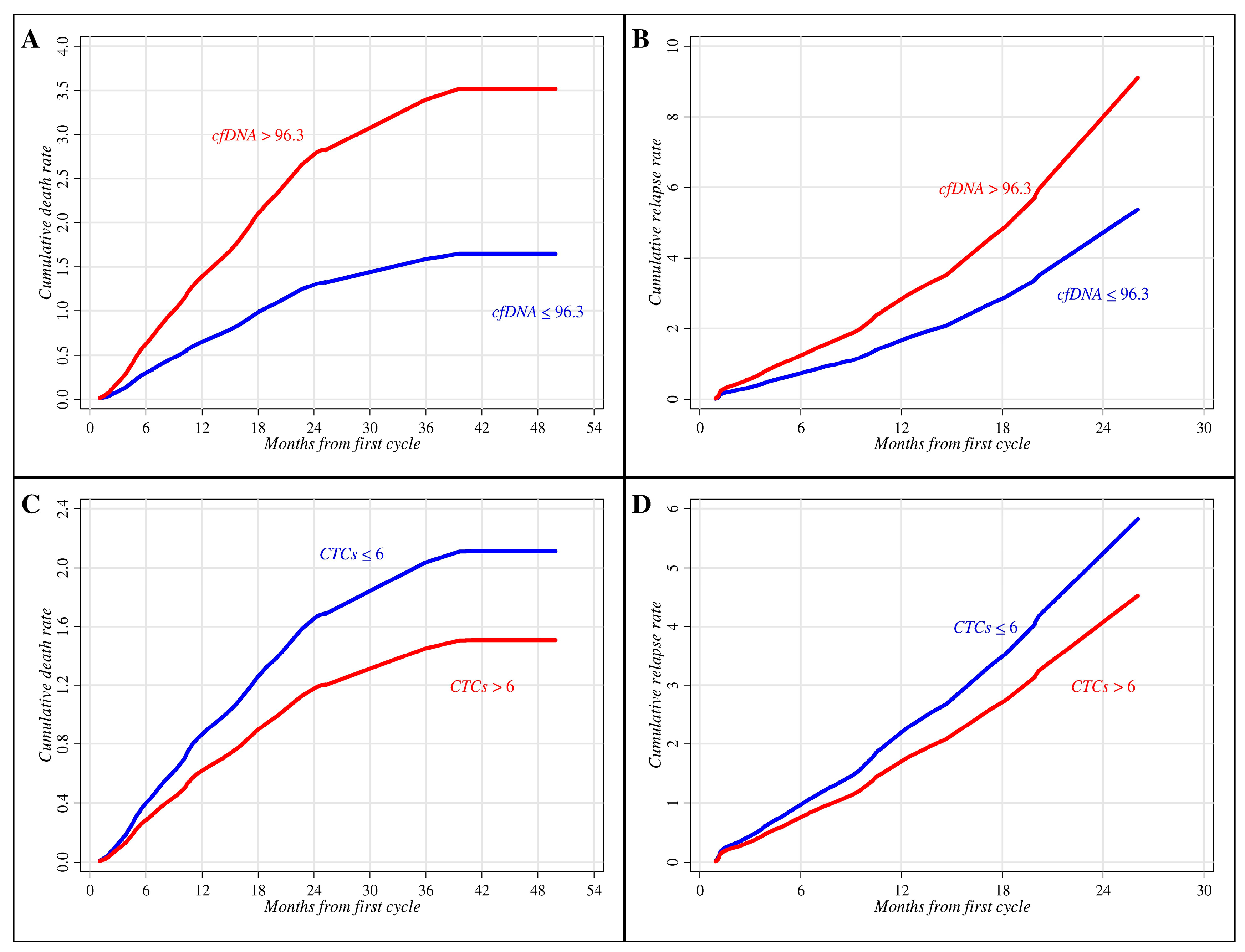
| ≤96.3 | 37 | 31 (83.8) | 0.38 | 0.23–0.53 | 0.019 |
| >96.3 | 35 | 33 (94.3) | 0.09 | 0.02–0.21 | |
| Not evaluable | 1 | 1 (100.0) | - | - | |
| Baseline CTCs | |||||
| ≤6 | 39 | 34 (87.2) | 0.18 | 0.08–0.31 | 0.402 |
| >6 | 34 | 31 (91.2) | 0.29 | 0.15–0.45 | |
| Age at first cycle | |||||
| ≤67 years | 37 | 31 (83.8) | 0.32 | 0.18–0.48 | 0.050 |
| >67 years | 36 | 34 (94.4) | 0.14 | 0.05–0.27 | |
| Gender | |||||
| Male | 50 | 44 (55.0) | 0.24 | 0.13–0.36 | 0.589 |
| Female | 23 | 22 (95.7) | 0.22 | 0.08–0.40 | |
| Histology | |||||
| Adenocarcinoma | 59 | 52 (88.1) | 0.24 | 0.14–0.35 | 0.546 |
| Squamous cell carcinoma | 14 | 13 (92.9) | 0.21 | 0.05–0.45 | |
| Metastases | |||||
| M1a | 23 | 22 (95.6%) | 0.22 | 0.08–0.40 | 0.763 |
| M1b | 50 | 43 (86.0%) | 0.24 | 0.13–0.36 | |
| Evidence of Brain Metastases | |||||
| Yes | 19 | 18 (94.7) | 0.16 | 0.04–0.35 | 0.243 |
| No | 54 | 47 (87.0) | 0.26 | 0.15–0.38 | |
| ECOG PS | |||||
| 0 | 21 | 16 (76.2) | 0.29 | 0.12–0.48 | 0.202 |
| 1–2 | 52 | 49 (94.2) | 0.21 | 0.11–0.33 | |
| Whole sample | 73 | 65 (89.0) | 0.23 | 0.14–0.34 | |
| Factors & Levels | Separate Effect | |||||
| PFS | OS | |||||
| HR | 95% CL | p-Value | HR | 95% CL | p-Value | |
| Baseline cfDNA | 0.040 | 0.006 | ||||
| ≤96.3 | 1.00 | 1.00 | ||||
| >96.3 | 1.70 | 1.02–2.83 | 2.14 | 1.24–3.68 | ||
| Baseline CTCs | 0.256 | 0.158 | ||||
| ≤6 | 1.00 | 1.00 | ||||
| >6 | 0.75 | 0.46–1.23 | 0.68 | 0.40–1.16 | ||
| Factors & Levels | Joint Effect | |||||
| PFS | OS | |||||
| HR | 95% CL | p-Value | HR | 95% CL | p-Value | |
| Baseline cfDNA | 0.075 | 0.012 | ||||
| ≤96.3 | 1.00 | 1.00 | ||||
| >96.3 | 1.63 | 0.95–2.78 | 2.03 | 1.17-3.52 | ||
| Baseline CTCs | 0.629 | 0.385 | ||||
| ≤6 | 1.00 | 1.00 | ||||
| >6 | 0.88 | 0.52–1.48 | 0.79 | 0.45–1.36 | ||
| Factors & Levels | Separate Effect | Joint Effect | ||||
|---|---|---|---|---|---|---|
| OS | OS | |||||
| HR | 95% CL | p-Value | HR | 95% CL | p-Value | |
| Baseline cfDNA | 0.047 | 0.153 | ||||
| ≤96.3 | 1.00 | 1.00 | ||||
| > 96.3 | 2.32 | 1.01–2.53 | 1.87 | 0.79–4.46 | ||
| Baseline CTC | 0.058 | 0.194 | ||||
| ≤6 | 1.00 | 1.00 | ||||
| >6 | 0.47 | 0.22–1.03 | 0.59 | 0.26–1.32 | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coco, S.; Alama, A.; Vanni, I.; Fontana, V.; Genova, C.; Dal Bello, M.G.; Truini, A.; Rijavec, E.; Biello, F.; Sini, C.; et al. Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy. Int. J. Mol. Sci. 2017, 18, 1035. https://doi.org/10.3390/ijms18051035
Coco S, Alama A, Vanni I, Fontana V, Genova C, Dal Bello MG, Truini A, Rijavec E, Biello F, Sini C, et al. Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy. International Journal of Molecular Sciences. 2017; 18(5):1035. https://doi.org/10.3390/ijms18051035
Chicago/Turabian StyleCoco, Simona, Angela Alama, Irene Vanni, Vincenzo Fontana, Carlo Genova, Maria Giovanna Dal Bello, Anna Truini, Erika Rijavec, Federica Biello, Claudio Sini, and et al. 2017. "Circulating Cell-Free DNA and Circulating Tumor Cells as Prognostic and Predictive Biomarkers in Advanced Non-Small Cell Lung Cancer Patients Treated with First-Line Chemotherapy" International Journal of Molecular Sciences 18, no. 5: 1035. https://doi.org/10.3390/ijms18051035






