Specific MicroRNA Pattern in Colon Tissue of Young Children with Eosinophilic Colitis
Abstract
:1. Introduction
2. Results
2.1. EC-Specific miR Expression Profile by Next-Generation Sequencing (NGS)
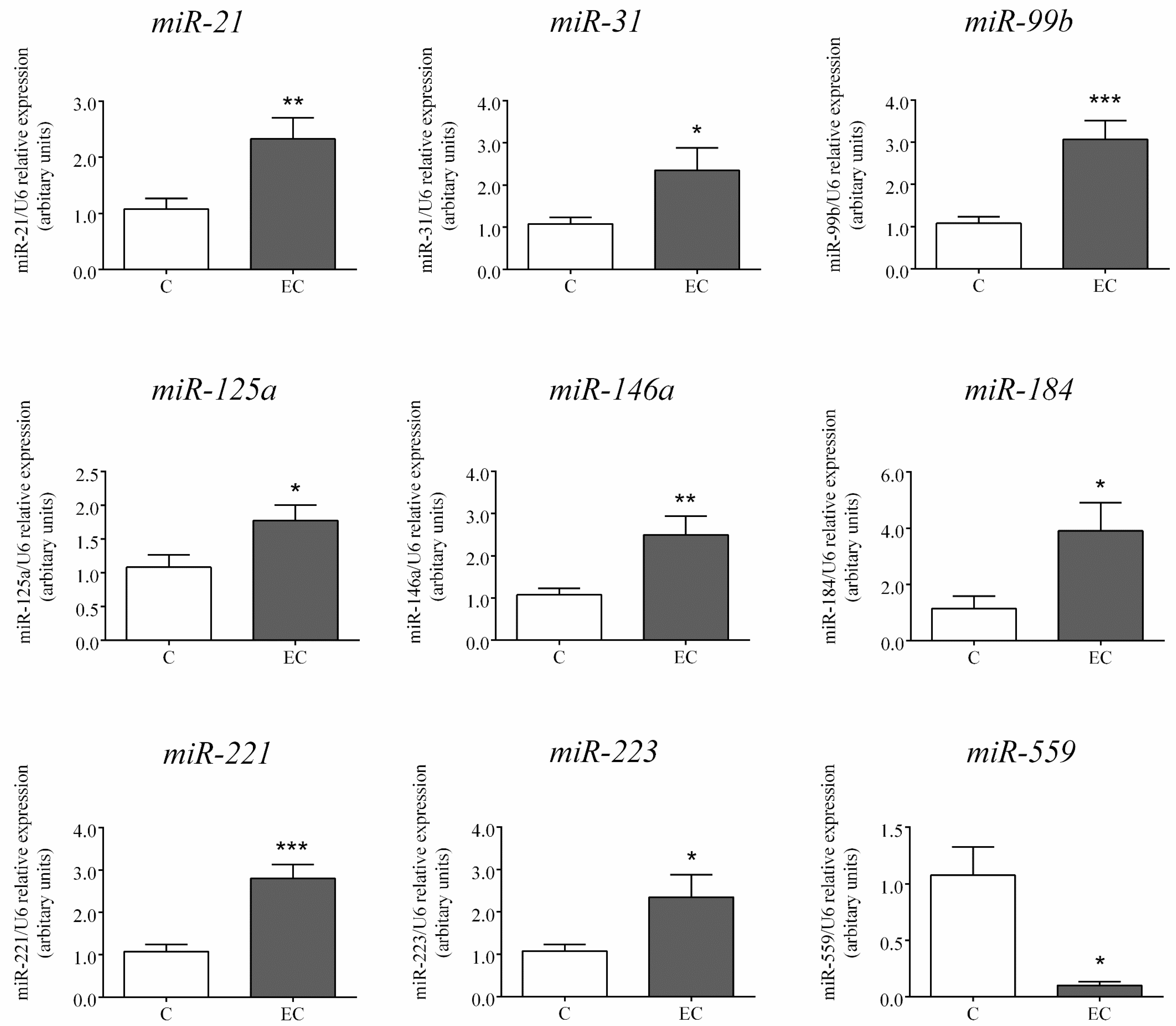
2.2. Expression of miRs Selected for Further Analysis by RT-PCR
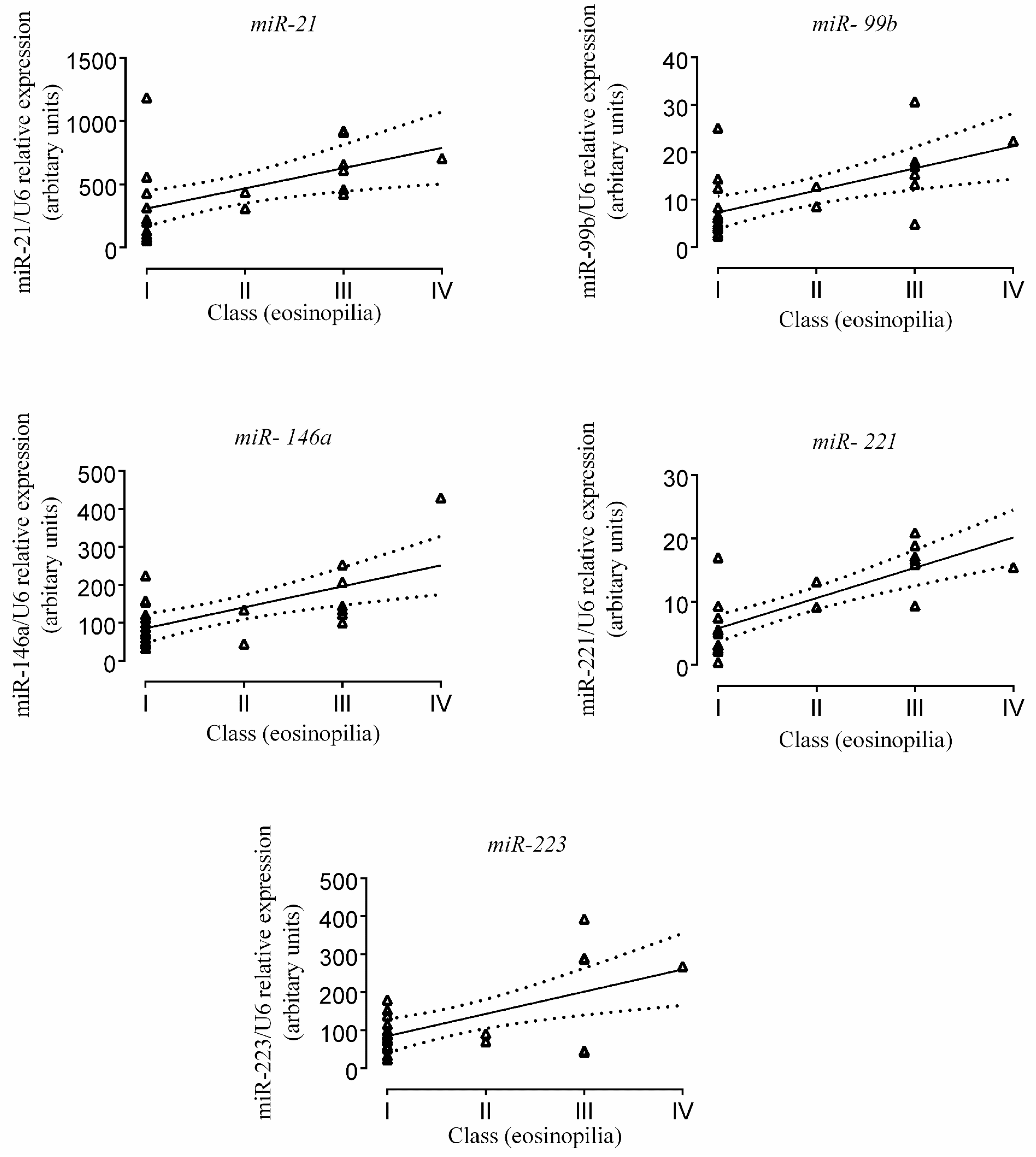
2.3. Correlation of miR Expression with Tissue Eosinophil Counts
2.4. Results of Bioinformatics Analysis
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. RNA Isolation
4.3. cDNA Library Preparation and NGS
4.4. RT-PCR
4.5. Bioinformatics Analysis of the Whole NGS miR Profile
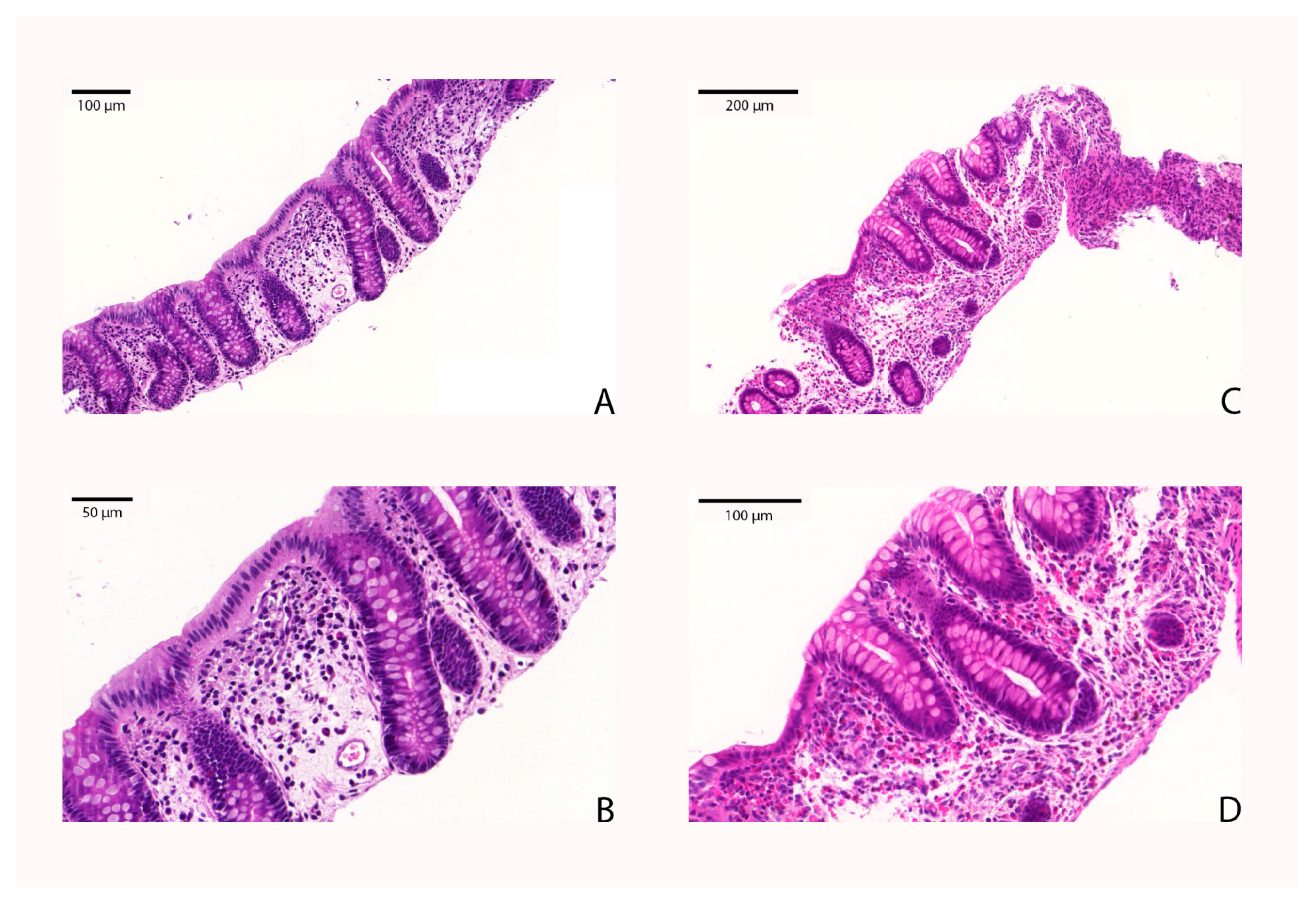
4.6. Determination of Eosinophil Counts in the Colonic Tissue of EC Patients
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fox, V.L. Gastrointestinal bleeding in infancy and childhood. Gastroenterol. Clin. N. Am. 2000, 29, 37–66. [Google Scholar] [CrossRef]
- Machida, H.M.; Catto Smith, A.G.; Gall, D.G.; Trevenen, C.; Scott, R.B. Allergic colitis in infancy: Clinical and pathologic aspects. J. Pediatr. Gastroenterol. Nutr. 1994, 19, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, M.E.; Mishra, A.; Brandt, E.B.; Hogan, S.P. Gastrointestinal eosinophils. Immunol. Rev. 2001, 179, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Alfadda, A.A.; Storr, M.A.; Shaffer, E.A. Eosinophilic colitis: An update on pathophysiology and treatment. Br. Med. Bull. 2011, 100, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Uppal, V.; Kreiger, P.; Kutsch, E. Eosinophilic gastroenteritis and colitis: A comprehensive review. Clin. Rev. Allergy Immunol. 2016, 50, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, Y.; Jimbo, K.; Inage, E.; Mori, M.; Yamakawa, Y.; Aoyagi, Y.; Suzuki, M.; Kudo, T.; Suzuki, R.; Shimizu, T. Microarray analysis of mucosal biopsy specimens in neonates with rectal bleeding: Is it really an allergic disease? J. Allergy Clin. Immunol. 2012, 129, 1676–1678. [Google Scholar] [CrossRef] [PubMed]
- Cseh, A.; Molnar, K.; Pinter, P.; Szalay, B.; Szebeni, B.; Treszl, A.; Arato, A.; Vasarhelyi, B.; Veres, G. Regulatory T cells and T helper subsets in breast-fed infants with hematochezia caused by allergic colitis. J. Pediatr. Gastroenterol. Nutr. 2010, 51, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Torrente, F.; Barabino, A.; Bellini, T.; Murch, S.H. Intraepithelial lymphocyte eotaxin-2 expression and perineural mast cell degranulation differentiate allergic/eosinophilic colitis from classic IBD. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.T.; Martin, C.F.; Kappelman, M.D.; Dellon, E.S. Prevalence of eosinophilic gastritis, gastroenteritis, and colitis: Estimates from a national administrative database. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Molnar, K.; Pinter, P.; Gyorffy, H.; Cseh, A.; Muller, K.E.; Arato, A.; Veres, G. Characteristics of allergic colitis in breast-fed infants in the absence of cow’s milk allergy. World J. Gastroenterol. 2013, 19, 3824–3830. [Google Scholar] [CrossRef] [PubMed]
- Cannioto, Z.; Berti, I.; Martelossi, S.; Bruno, I.; Giurici, N.; Crovella, S.; Ventura, A. IBD and IBD mimicking enterocolitis in children younger than 2 years of age. Eur. J. Pediatr. 2009, 168, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Katsanos, K.H.; Zinovieva, E.; Lambri, E.; Tsianos, E.V. Eosinophilic–Crohn overlap colitis and review of the literature. J. Crohns Colitis 2011, 5, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Bala, G.; Swincow, G.; Rytarowska, A.; Czerwionka-Szaflarska, M. Non-specific inflammatory bowel disease or allergic colitis in small children—What is the final diagnosis. Med. Wieku Rozwoj. 2006, 10, 475–482. [Google Scholar] [PubMed]
- Ozen, A.; Gulcan, E.M.; Saricoban, H.E.; Ozkan, F.; Cengizlier, R. Food protein-induced non-immunoglobulin E-mediated allergic colitis in infants and older children: What cytokines are involved? Int. Arch. Allergy Immunol. 2015, 168, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, S.C. Food allergy and eosinophilic gastroenteritis and colitis. Curr. Opin. Allergy Clin. Immunol. 2010, 10, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Kalla, R.; Ventham, N.T.; Kennedy, N.A.; Quintana, J.F.; Nimmo, E.R.; Buck, A.H.; Satsangi, J. MicroRNAs: New players in IBD. Gut 2015, 64, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Wang, J.Y. RNA-binding proteins and microRNAs in gastrointestinal epithelial homeostasis and diseases. Curr. Opin. Pharmacol. 2014, 19, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Zahm, A.M.; Menard-Katcher, C.; Benitez, A.J.; Tsoucas, D.M.; le Guen, C.L.; Hand, N.J.; Friedman, J.R. Pediatric eosinophilic esophagitis is associated with changes in esophageal microRNAs. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G803–G812. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Sherrill, J.D.; Wen, T.; Plassard, A.J.; Besse, J.A.; Abonia, J.P.; Franciosi, J.P.; Putnam, P.E.; Eby, M.; Martin, L.J.; et al. MicroRNA signature in patients with eosinophilic esophagitis, reversibility with glucocorticoids, and assessment as disease biomarkers. J. Allergy Clin. Immunol. 2012, 129, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- Pua, H.H.; Ansel, K.M. MicroRNA regulation of allergic inflammation and asthma. Curr. Opin. Immunol. 2015, 36, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Munitz, A.; Rothenberg, M.E. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 2009, 182, 4994–5002. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2α. Gastroenterology 2008, 135, 1624–1635. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Lim, E.J.; Itskovich, S.; Besse, J.A.; Plassard, A.J.; Mingler, M.K.; Rothenberg, J.A.; Fulkerson, P.C.; Aronow, B.J.; Rothenberg, M.E. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS ONE 2013, 8, e59397. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Lau, K.M.; Chan, I.H.; Hu, S.; Lam, Y.Y.; Choi, A.O.; Lam, C.W. MicroRNA-21* regulates the prosurvival effect of GM-CSF on human eosinophils. Immunobiology 2013, 218, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Hartner, J.; Lim, E.J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-γ pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef] [PubMed]
- Hennigan, K.; Conroy, P.J.; Walsh, M.T.; Amin, M.; O’Kennedy, R.; Ramasamy, P.; Gleich, G.J.; Siddiqui, Z.; Glynn, S.; McCabe, O.; et al. Eosinophil peroxidase activates cells by HER2 receptor engagement and β1-integrin clustering with downstream MAPK cell signaling. Clin. Immunol. 2016, 171, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, B.P.; Olsen, P.C.; Coelho, L.P.; Ferreira, T.P.; Souza, H.S.; Martins, M.A.; Viola, J.P. NFAT1 transcription factor regulates pulmonary allergic inflammation and airway responsiveness. Am. J. Respir. Cell Mol. Biol. 2009, 40, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Weitzel, R.P.; Lesniewski, M.L.; Haviernik, P.; Kadereit, S.; Leahy, P.; Greco, N.J.; Laughlin, M.J. 184 regulates expression of NFAT1 in umbilical cord blood CD4+ T cells. Blood 2009, 113, 6648–6657. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hui, Y.; Xing, C.; Guo, Y.; Wang, Q.; Shu, J.; Qian, J.; Zhou, G. MicroRNA-31 affects the expression of asthma-related cytokines via regulation of CD44. Int. J. Clin. Exp. Med. 2016, 9, 11. [Google Scholar]
- Abdulnour-Nakhoul, S.M.; Al-Tawil, Y.; Gyftopoulos, A.A.; Brown, K.L.; Hansen, M.; Butcher, K.F.; Eidelwein, A.P.; Noel, R.A.; Rabon, E.; Posta, A.; et al. Alterations in junctional proteins, inflammatory mediators and extracellular matrix molecules in eosinophilic esophagitis. Clin. Immunol. 2013, 148, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Cook, D.; Walker, M.M.; Talley, N.J. Pharmacological treatment of eosinophilic gastrointestinal disorders. Expert Rev. Clin. Pharmacol. 2016, 9, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.; Li, Y.; Kang, M. Activation of miR-21 by STAT3 induces proliferation and suppresses apoptosis in nasopharyngeal carcinoma by targeting PTEN gene. PLoS ONE 2014, 9, e109929. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Tymen, S.D.; Chen, D.; Fang, Z.J.; Zhao, Y.; Dragas, D.; Dai, Y.; Marucha, P.T.; Zhou, X. MicroRNA-99 family targets AKT/mTOR signaling pathway in dermal wound healing. PLoS ONE 2013, 8, e64434. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Kang, N.; He, L.; Li, X.; Xu, X.; Zhang, H. MiR-221 and miR-26b regulate chemotactic migration of MSCs toward HGF through activation of Akt and FAK. J. Cell. Biochem. 2016, 117, 1370–1383. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Mukaisyo, K.; Sugihara, H.; Fujiyama, Y.; Hattori, T. Increased apoptosis and decreased proliferation of colonic epithelium in dextran sulfate sodium-induced colitis in mice. Oncol. Rep. 2010, 24, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Lou, Y.; Porturas, T.; Morrissey, S.; Luo, G.; Qi, J.; Ruan, Q.; Shi, S.; Chen, Y.H. Exacerbated experimental colitis in TNFAIP8-deficient mice. J. Immunol. 2015, 194, 5736–5742. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Li, Y.C.; Toback, F.G. AMP-18 Targets p21 to maintain epithelial homeostasis. PLoS ONE 2015, 10, e0125490. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, H.; Masuda, T.; Maisawa, S.; Chida, S. Apoptotic epithelial cells in biopsy specimens from infants with streaked rectal bleeding. J. Pediatr. Gastroenterol. Nutr. 2001, 32, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Eyers, F.; Xiang, Y.; Guo, M.; Young, I.G.; Rosenberg, H.F.; Foster, P.S. Expression profiling of differentiating eosinophils in bone marrow cultures predicts functional links between microRNAs and their target mRNAs. PLoS ONE 2014, 9, e97537. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yang, Q.; Xu, H.; Zhang, J.; Deng, H.; Gao, H.; Yang, J.; Zhao, D.; Liu, F. miRNA-221-3p Enhances the secretion of interleukin-4 in mast cells through the phosphatase and tensin homolog/p38/nuclear factor-κB pathway. PLoS ONE 2016, 11, e0148821. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Lim, E.J.; Besse, J.A.; Itskovich, S.; Plassard, A.J.; Fulkerson, P.C.; Aronow, B.J.; Rothenberg, M.E. MiR-223 deficiency increases eosinophil progenitor proliferation. J. Immunol. 2013, 190, 1576–1582. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.J.; Ngeow, J.; Collins, M.H.; Martin, L.J.; Putnam, P.E.; Abonia, J.P.; Marsolo, K.; Eng, C.; Rothenberg, M.E. Increased prevalence of eosinophilic gastrointestinal disorders in pediatric PTEN hamartoma tumor syndromes. J. Pediatr. Gastroenterol. Nutr. 2014, 58, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Gui, F.; Hong, Z.; You, Z.; Wu, H.; Zhang, Y. MiR-21 inhibitor suppressed the progression of retinoblastoma via the modulation of PTEN/PI3K/AKT pathway. Cell Biol. Int. 2016, 40, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Liu, H.; Xia, L.X.; Tian, B.P.; Huang, H.Q.; Chen, Z.Y.; Ju, Z.Y.; Li, W.; Chen, Z.H.; Shen, H.H. Rapamycin inhibition of eosinophil differentiation attenuates allergic airway inflammation in mice. Respirology 2015, 20, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Cao, A.; Yao, S.; Evans-Marin, H.L.; Liu, H.; Wu, W.; Carlsen, E.D.; Dann, S.M.; Soong, L.; Sun, J.; Zhao, Q.; Cong, Y. mTOR Mediates IL-23 induction of neutrophil IL-17 and IL-22 production. J. Immunol. 2016, 196, 4390–4399. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Takeda, M.; Nishikawa, J.; Konno, Y.; Tamaki, M.; Itoga, M.; Kobayashi, Y.; Moritoki, Y.; Ito, W.; et al. The effect of pharmacological PI3Kγ inhibitor on eotaxin-induced human eosinophil functions. Pulm. Pharmacol. Ther. 2014, 27, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- UniProt Consortium. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar]


| A | Control | Eosinophilic Colitis |
| n | 4 | 4 |
| Male/Female | 2/2 | 3/1 |
| Age (months) | 136.5 ± 35.78 | 8.88 ± 5.14 * |
| BMI (kg/m2) | - | 18.66 ± 3.79 |
| Iron (µmol/L) | 10.75 ± 2.5 | 13.25 ± 3.57 |
| Albumin (g/L) | 51.75 ± 8.81 | - |
| Haemoglobin (g/L) | 134.3 ± 7.76 | 121.8 ± 3.64 |
| Haematocrit (%) | 0.39 ± 0.02 | 0.34 ± 0.013 |
| Platelet count (Giga/L) | 325.8 ± 21.10 | 440.5 ± 57.81 |
| CRP (mg/L) | 1.25 ± 1.4 | 0 |
| B | Control | Eosinophilic Colitis |
| n | 14 | 10 |
| Male/Female | 6/8 | 8/2 |
| Age (months) | 29.82 ± 17.08 | 5.7 ± 4.76 *** |
| BMI (kg/m2) | 16.15 ± 1.6 | 16.21 ± 1.74 |
| Length-weight percentile (%) | - | 42.25 ± 34.92 1/10 overweight 1/10 underweight |
| Iron (µmol/L) | 15.70 ± 4.9 | 9.89 ± 5.62 * |
| Albumin (g/L) | 45.8 ± 2.01 | 44 ± 4.24 |
| Haemoglobin (g/L) | 122.1 ± 12.26 | 112.6 ± 8.32 * |
| Haematocrit (%) | 35.18 ± 2.8 | 32 ± 1.94 |
| Platelet count (Giga/L) | 364.6 ± 87.09 | 394.5 ± 104.1 |
| CRP (mg/L) | 0.79 ± 1.4 | 3.44 ± 8.49 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kiss, Z.; Béres, N.J.; Sziksz, E.; Tél, B.; Borka, K.; Arató, A.; Szabó, A.J.; Veres, G. Specific MicroRNA Pattern in Colon Tissue of Young Children with Eosinophilic Colitis. Int. J. Mol. Sci. 2017, 18, 1050. https://doi.org/10.3390/ijms18051050
Kiss Z, Béres NJ, Sziksz E, Tél B, Borka K, Arató A, Szabó AJ, Veres G. Specific MicroRNA Pattern in Colon Tissue of Young Children with Eosinophilic Colitis. International Journal of Molecular Sciences. 2017; 18(5):1050. https://doi.org/10.3390/ijms18051050
Chicago/Turabian StyleKiss, Zoltán, Nóra Judit Béres, Erna Sziksz, Bálint Tél, Katalin Borka, András Arató, Attila J. Szabó, and Gábor Veres. 2017. "Specific MicroRNA Pattern in Colon Tissue of Young Children with Eosinophilic Colitis" International Journal of Molecular Sciences 18, no. 5: 1050. https://doi.org/10.3390/ijms18051050






