dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster
Abstract
:1. Introduction
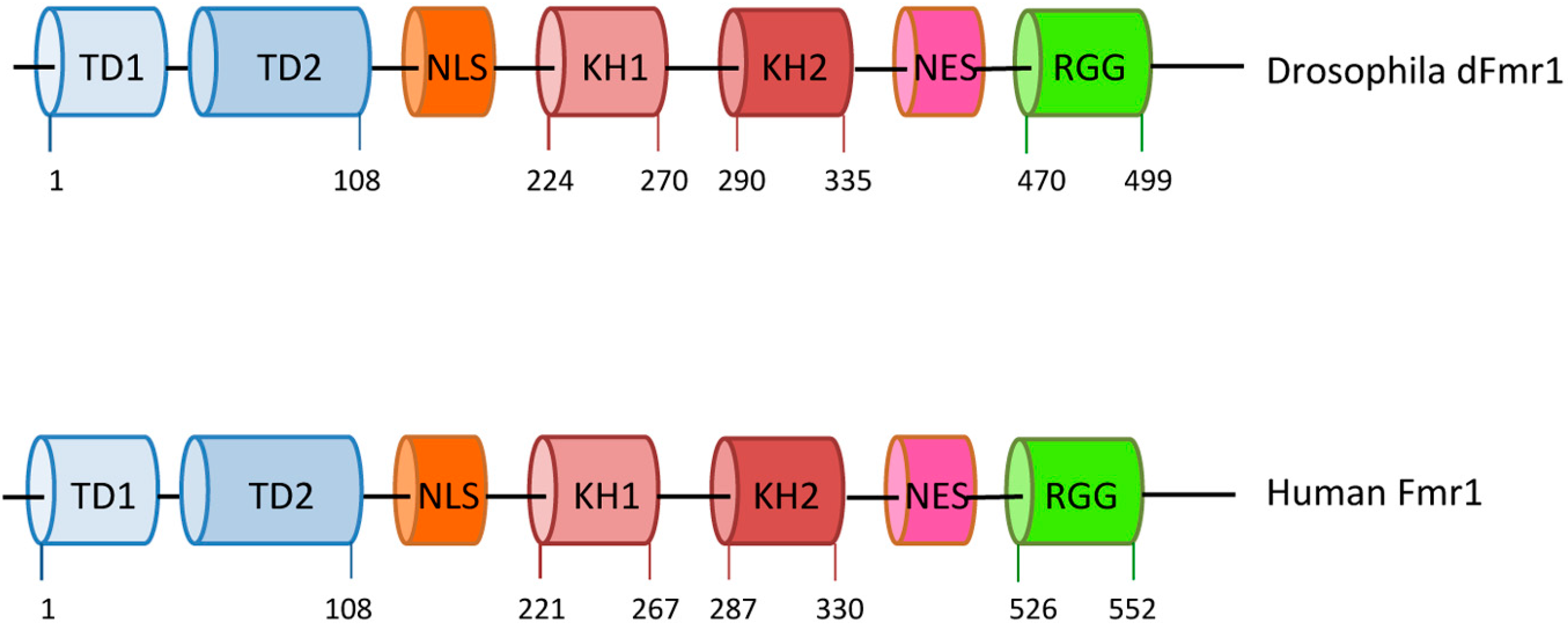
1.1. The Fragile-X Mental Retardation Gene
1.2. Drosophila Small RNA Pathways: An Overview
1.3. piRNA Genes Are Conserved during Evolution
2. The piRNA Pathway in Drosophila Gonads
2.1. The piRNA Pathway in the Ovary
2.2. The piRNA Pathway in the Testes and Its Role in the crystal-Stellate Regulation
3. dFmr1 Participates in the piRNA Pathway Occurring in Gonads
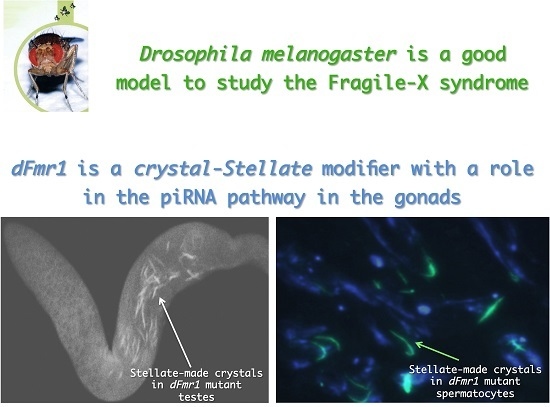
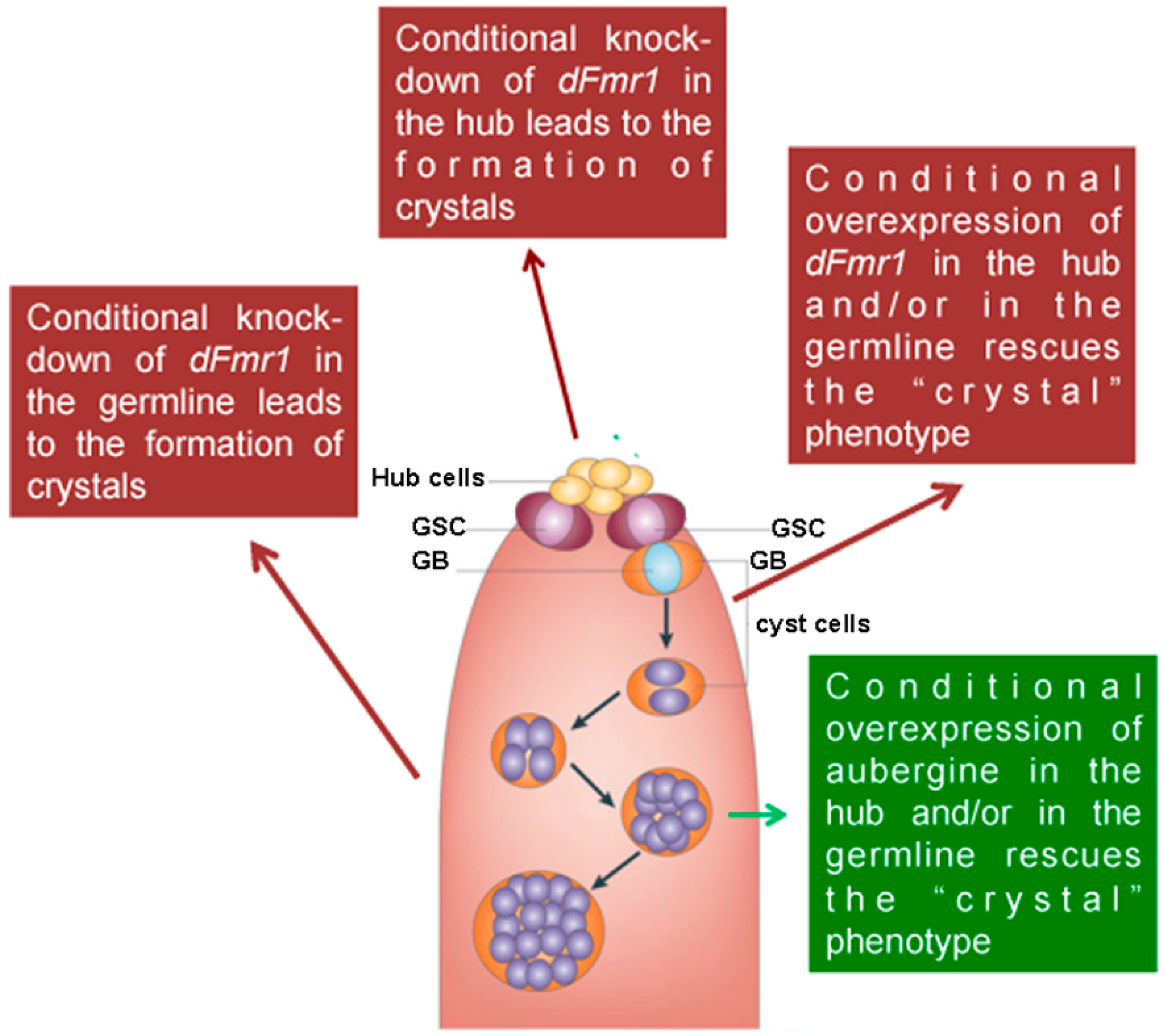
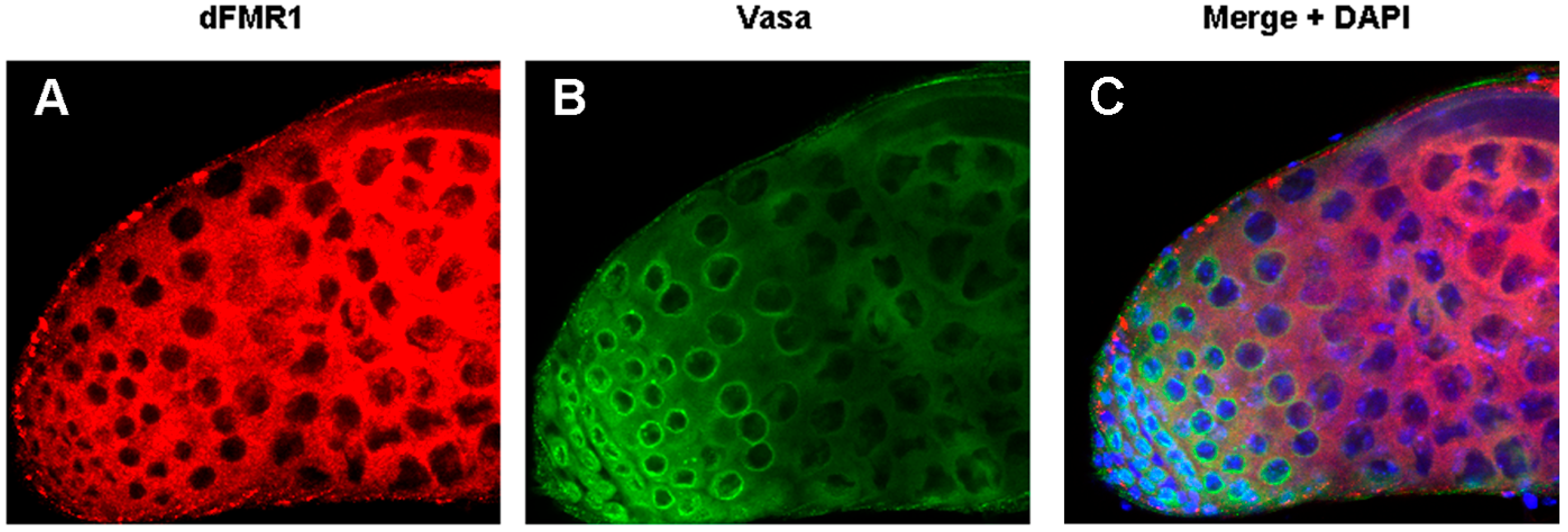
3.1. dFmr1’s Role in the Testes
3.2. dFmr1’s Role in the Ovary
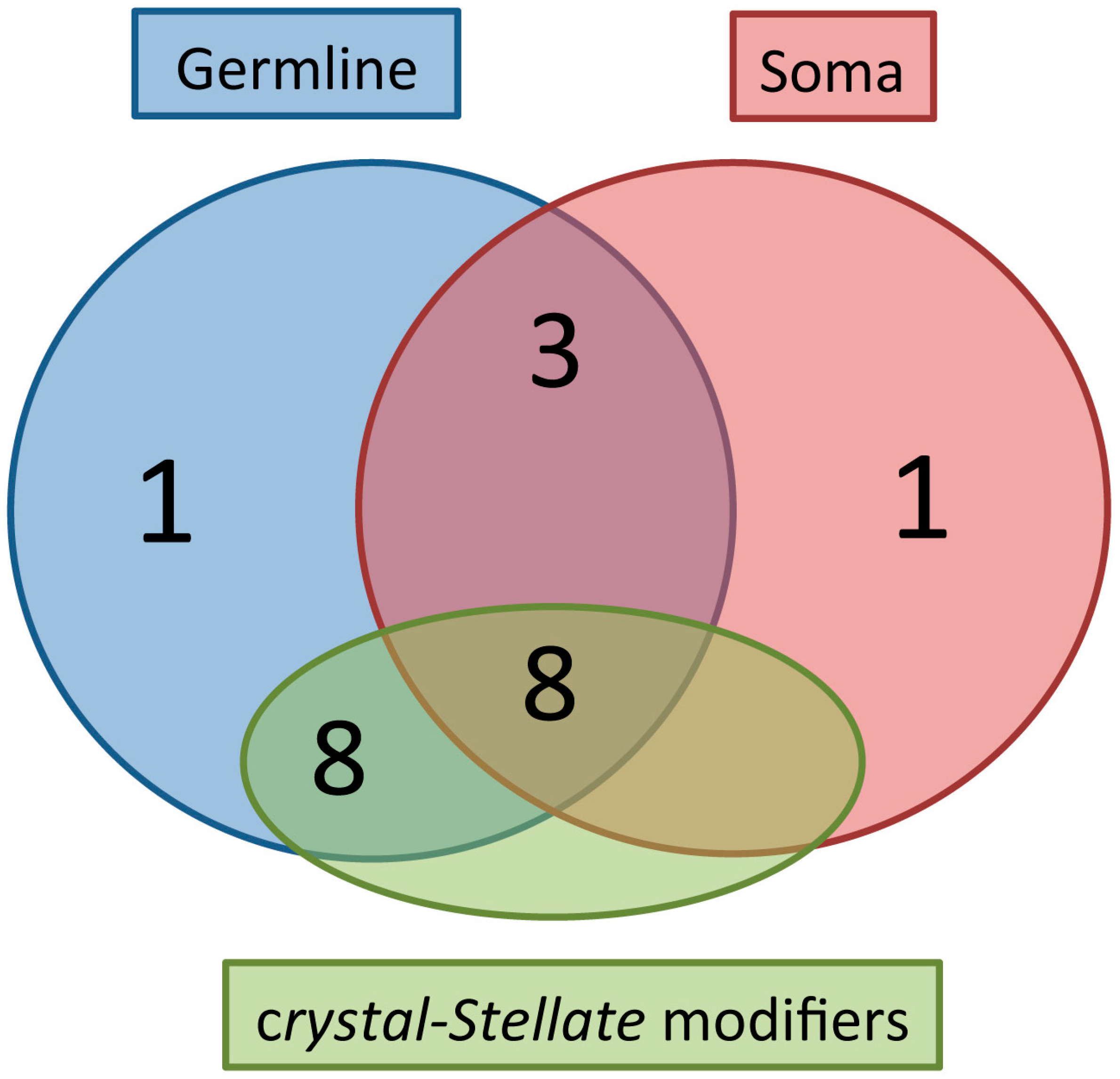
4. Genetic and Biochemical Interactors of dFmr1
Nuclear Interactors in the Small RNA Pathway (in the Gonads)
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Siomi, M.C.; Siomi, H.; Sauer, W.H.; Srinivasan, S.; Nussbaum, R.L.; Dreyfuss, G. FXR1, an autosomal homolog of the fragile X mental retardation gene. EMBO J. 1995, 14, 2401–2408. [Google Scholar] [PubMed]
- Zhang, Y.; O’Connor, J.P.; Siomi, M.C.; Srinivasan, S.; Dutra, A.; Nussbaum, R.L.; Dreyfuss, G. The fragile X mental retardation syndrome protein interacts with novel homologs FXR1 and FXR2. EMBO J. 1995, 14, 5358–5366. [Google Scholar] [PubMed]
- Adams-Cioaba, M.A.; Guo, Y.; Bian, C.; Amaya, M.F.; Lam, R.; Wasney, G.A.; Vedadi, M.; Xu, C.; Min, J. Structural studies of the tandem Tudor domains of fragile X mental retardation related proteins FXR1 and FXR2. PLoS ONE 2010, 5, e13559. [Google Scholar] [CrossRef] [PubMed]
- Schenck, A.; Bardoni, B.; Langmann, C.; Harden, N.; Mandel, J.L.; Giangrande, A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron 2003, 38, 887–898. [Google Scholar] [CrossRef]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; de Rubeis, S.; di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.E.; Lorsch, J.R. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 2012, 19, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Caudy, A.A.; Myers, M.; Hannon, G.J.; Hammond, S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002, 16, 2491–2496. [Google Scholar] [CrossRef] [PubMed]
- Ishizuka, A.; Siomi, M.C.; Siomi, H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002, 16, 2497–2508. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Zarnescu, D.C.; Ceman, S.; Nakamoto, M.; Mowrey, J.; Jongens, T.A.; Nelson, D.L.; Moses, K.; Warren, S.T. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 2004, 7, 113–117. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Antion, M.D.; Hu, D.; Spencer, C.M.; Paylor, R.; Klann, E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron 2006, 51, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Li, Y.; Wang, F.; Gao, F.B. The steady-state level of the nervous-system-specific microRNA-124a is regulated by dFMR1 in Drosophila. J. Neurosci. 2008, 28, 11883–11889. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Zhang, Y.; Siomi, H.; Dreyfuss, G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 1996, 16, 3825–3832. [Google Scholar] [CrossRef] [PubMed]
- Tamanini, F.; Meijer, N.; Verheij, C.; Willems, P.J.; Galjaard, H.; Oostra, B.A.; Hoogeveen, A.T. FMRP is associated to the ribosomes via RNA. Hum. Mol. Genet. 1996, 5, 809–813. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Absher, D.; Eberhart, D.E.; Brown, V.; Malter, H.E.; Warren, S.T. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1997, 1, 109–118. [Google Scholar] [CrossRef]
- Darnell, J.C.; Fraser, C.E.; Mostovetsky, O.; Stefani, G.; Jones, T.A.; Eddy, S.R.; Darnell, R.B. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005, 19, 903–918. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.R.; Bray, S.M.; Warren, S.T. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu. Rev. Pathol. 2012, 7, 219–245. [Google Scholar] [CrossRef] [PubMed]
- Zarnescu, D.C.; Jin, P.; Betschinger, J.; Nakamoto, M.; Wang, Y.; Dockendorff, T.C.; Feng, Y.; Jongens, T.A.; Sisson, J.C.; Knoblich, J.A.; et al. Fragile X protein functions with Lgl and the PAR complex in flies and mice. Dev. Cell 2005, 8, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Dockendorff, T.C.; Jongens, T.A.; Dreyfuss, G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol. Cell. Biol. 2000, 20, 8536–8547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Bailey, A.M.; Matthies, H.J.; Renden, R.B.; Smith, M.A.; Speese, S.D.; Rubin, G.M.; Broadie, K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell 2001, 107, 591–603. [Google Scholar] [CrossRef]
- Zhang, Y.Q.; Matthies, H.J.; Mancuso, J.; Andrews, H.K.; Woodruff, E., 3rd; Friedman, D.; Broadie, K. The Drosophila fragile X-related gene regulates axoneme differentiation during spermatogenesis. Dev. Biol. 2004, 270, 290–307. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Joseph, S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie 2015, 114, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Choi, M.; Siomi, M.C.; Nussbaum, R.L.; Dreyfuss, G. Essential role for KH domains in RNA binding: Impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell 1994, 77, 33–39. [Google Scholar] [CrossRef]
- Siomi, H.; Dreyfuss, G. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 1995, 129, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Bardoni, B.; Sittler, A.; Shen, Y.; Mandel, J.L. Analysis of domains affecting intracellular localization of the FMRP protein. Neurobiol. Dis. 1997, 4, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, M.; Zamore, P.D. Small silencing RNAs: An expanding universe. Nat. Rev. Genet. 2009, 10, 94–108. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Agbu, P.; Giri, R. MicroRNA function in Drosophila melanogaster. Semin. Cell Dev. Biol. 2016, 66, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Senti, K.A.; Brennecke, J. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet. 2010, 26, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-interacting RNA: Its biogenesis and functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of piRNAs in the central nervous system. RNA 2011, 17, 1090–1099. [Google Scholar] [CrossRef] [PubMed]
- Perrat, P.N.; DasGupta, S.; Wang, J.; Theurkauf, W.; Weng, Z.; Rosbash, M.; Waddell, S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013, 340, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Thomson, T.; Lin, H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu. Rev. Cell Dev. Biol. 2009, 25, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Forstemann, K.; Horwich, M.D.; Wee, L.; Tomari, Y.; Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 2007, 130, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Benna, C.; Mazzotta, G.M.; Piccin, A.; Zordan, M.A.; Costa, R.; Bozzetti, M.P. aubergine gene overexpression in somatic tissues of aubergine(sting) mutants interferes with the RNAi pathway of a yellow hairpin dsRNA in Drosophila melanogaster. Genetics 2008, 178, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R.; Schnabl, J.; Handler, D.; Mohn, F.; Ameres, S.L.; Brennecke, J. Genetic and mechanistic diversity of piRNA 3′-end formation. Nature 2016, 539, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Golden, R.J.; Chen, B.; Li, T.; Braun, J.; Manjunath, H.; Chen, X.; Wu, J.; Schmid, V.; Chang, T.C.; Kopp, F.; et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 2017, 542, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Lo, P.K.; Huang, Y.C.; Poulton, J.S.; Leake, N.; Palmer, W.H.; Vera, D.; Xie, G.; Klusza, S.; Deng, W.M. RNA helicase Belle/DDX3 regulates transgene expression in Drosophila. Dev. Biol. 2016, 412, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lin, H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2002, 2, 819–830. [Google Scholar] [CrossRef]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Reuter, M.; Chuma, S.; Tanaka, T.; Franz, T.; Stark, A.; Pillai, R.S. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009, 16, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Vourekas, A.; Zheng, K.; Fu, Q.; Maragkakis, M.; Alexiou, P.; Ma, J.; Pillai, R.S.; Mourelatos, Z.; Wang, P.J. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015, 29, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Soper, S.F.; van der Heijden, G.W.; Hardiman, T.C.; Goodheart, M.; Martin, S.L.; de Boer, P.; Bortvin, A. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev. Cell 2008, 15, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K.C.; Xing, J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genom. 2014, 15, 545. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, H.J.; Heyn, H.; Garcia del Muro, X.; Vidal, A.; Larriba, S.; Munoz, C.; Villanueva, A.; Esteller, M. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics 2014, 9, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Preall, J.B.; McGinn, J.; Hannon, G.J. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol. Cell 2013, 50, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Handler, D.; Meixner, K.; Pizka, M.; Lauss, K.; Schmied, C.; Gruber, F.S.; Brennecke, J. The genetic makeup of the Drosophila piRNA pathway. Mol. Cell 2013, 50, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Muerdter, F.; Guzzardo, P.M.; Gillis, J.; Luo, Y.; Yu, Y.; Chen, C.; Fekete, R.; Hannon, G.J. A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila. Mol. Cell 2013, 50, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Toth, K.F.; Aravin, A.A. To be or not to be a piRNA: Genomic origin and processing of piRNAs. Genome Biol. 2014, 15, 204. [Google Scholar] [CrossRef] [PubMed]
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.C.; Luo, Y.; Sachidanandam, R.; Toth, K.F.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Eddy, E.M. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat. Rec. 1974, 178, 731–757. [Google Scholar] [CrossRef] [PubMed]
- Eddy, E.M. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1975, 43, 229–280. [Google Scholar] [PubMed]
- Snee, M.J.; Macdonald, P.M. Live imaging of nuage and polar granules: Evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 2004, 117 Pt 10, 2109–2120. [Google Scholar] [CrossRef] [PubMed]
- Lim, A.K.; Kai, T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2007, 104, 6714–6719. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2008, 135, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Nagao, A.; Sato, K.; Nishida, K.M.; Siomi, H.; Siomi, M.C. Gender-Specific Hierarchy in Nuage Localization of PIWI-Interacting RNA Factors in Drosophila. Front. Genet. 2011, 2, 55. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Lehmann, R.; Teixeira, F.K. The cellular basis of hybrid dysgenesis and Stellate regulation in Drosophila. Curr. Opin. Genet. Dev. 2015, 34, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ishizu, H.; Saito, K.; Fukuhara, S.; Kamatani, M.K.; Bonnefond, L.; Matsumoto, N.; Nishizawa, T.; Nakanaga, K.; Aoki, J.; et al. Structure and function of Zucchini endoribonuclease in piRNA biogenesis. Nature 2012, 491, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Ipsaro, J.J.; Haase, A.D.; Knott, S.R.; Joshua-Tor, L.; Hannon, G.J. The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis. Nature 2012, 491, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Szakmary, A.; Reedy, M.; Qi, H.; Lin, H. The Yb protein defines a novel organelle and regulates male germline stem cell self-renewal in Drosophila melanogaster. J. Cell Biol. 2009, 185, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Haase, A.D.; Fenoglio, S.; Muerdter, F.; Guzzardo, P.M.; Czech, B.; Pappin, D.J.; Chen, C.; Gordon, A.; Hannon, G.J. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010, 24, 2499–2504. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, D.; Sykora, M.M.; Sachidanandam, R.; Mechtler, K.; Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010, 29, 3301–3317. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ishizu, H.; Komai, M.; Kotani, H.; Kawamura, Y.; Nishida, K.M.; Siomi, H.; Siomi, M.C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010, 24, 2493–2498. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Watanabe, T.; Ku, H.Y.; Liu, N.; Zhong, M.; Lin, H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 2011, 286, 3789–3797. [Google Scholar] [CrossRef] [PubMed]
- Dennis, C.; Zanni, V.; Brasset, E.; Eymery, A.; Zhang, L.; Mteirek, R.; Jensen, S.; Rong, Y.S.; Vaury, C. “Dot COM”, a nuclear transit center for the primary piRNA pathway in Drosophila. PLoS ONE 2013, 8, e72752. [Google Scholar] [CrossRef] [PubMed]
- Murota, Y.; Ishizu, H.; Nakagawa, S.; Iwasaki, Y.W.; Shibata, S.; Kamatani, M.K.; Saito, K.; Okano, H.; Siomi, H.; Siomi, M.C. Yb Integrates piRNA intermediates and processing factors into perinuclear bodies to enhance piRISC assembly. Cell Rep. 2014, 8, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, J.; Xu, J.; Zhang, Z.; Koppetsch, B.S.; Schultz, N.; Vreven, T.; Meignin, C.; Davis, I.; Zamore, P.D.; et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 2012, 151, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Sato, K.; Pezic, D.; Aravin, A.A. PIWI-interacting small RNAs: The vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011, 12, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.; Marconi, S.; Specchia, V.; Bozzetti, M.P.; Ivics, Z.; Caizzi, R.; Marsano, R.M. Functional characterization of the Bari1 transposition system. PLoS ONE 2013, 8, e79385. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pane, A.; Schupbach, T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr. Biol. 2007, 17, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Bratu, D.P.; McGinnis-Schultz, N.; Koppetsch, B.S.; Cook, H.A.; Theurkauf, W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 2007, 12, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Pane, A.; Wehr, K.; Schupbach, T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev. Cell 2007, 12, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.M.; Saito, K.; Mori, T.; Kawamura, Y.; Nagami-Okada, T.; Inagaki, S.; Siomi, H.; Siomi, M.C. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007, 13, 1911–1922. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Kai, T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 2010, 20, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, J.; Koppetsch, B.S.; Wang, J.; Tipping, C.; Ma, S.; Weng, Z.; Theurkauf, W.E.; Zamore, P.D. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell 2011, 44, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.; Cora, E.; Coute, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R.; et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.; Li, S.; Hur, J.K.; Wachsmuth, M.; Bois, J.S.; Perkins, E.M.; Patel, D.J.; Aravin, A.A. Aub and Ago3 are recruited to nuage through two mechanisms to form a ping-pong complex assembled by krimper. Mol. Cell 2015, 59, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Sahin, H.B.; Karatas, O.F.; Specchia, V.; Tommaso, S.D.; Diebold, C.; Bozzetti, M.P.; Giangrande, A. Novel mutants of the aubergine gene. Fly 2016, 10, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Iwasaki, Y.W.; Shibuya, A.; Carninci, P.; Tsuchizawa, Y.; Ishizu, H.; Siomi, M.C.; Siomi, H. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell 2015, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Gatti, M.; Pimpinelli, S.; Santini, G. Characterization of Drosophila heterochromatin. I. Staining and decondensation with Hoechst 33258 and quinacrine. Chromosoma 1976, 57, 351–375. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, G.; Bonaccorsi, S.; Robbins, L.G.; Pimpinelli, S. Genetic analysis of Stellate elements of Drosophila melanogaster. Genetics 1994, 138, 1181–1197. [Google Scholar] [PubMed]
- Livak, K.J. Organization and mapping of a sequence on the Drosophila melanogaster X and Y chromosomes that is transcribed during spermatogenesis. Genetics 1984, 107, 611–634. [Google Scholar] [PubMed]
- Livak, K.J. Detailed structure of the Drosophila melanogaster stellate genes and their transcripts. Genetics 1990, 124, 303–316. [Google Scholar] [PubMed]
- Shevelyov, Y.Y. Copies of a Stellate gene variant are located in the X heterochromatin of Drosophila melanogaster and are probably expressed. Genetics 1992, 132, 1033–1037. [Google Scholar] [PubMed]
- Bozzetti, M.P.; Fanti, L.; Di Tommaso, S.; Piacentini, L.; Berloco, M.; Tritto, P.; Specchia, V. The “Special” crystal-Stellate System in Drosophila melanogaster Reveals Mechanisms Underlying piRNA Pathway-Mediated Canalization. Genet. Res. Int. 2012, 2012, 5. [Google Scholar] [CrossRef]
- Meyer, G.F.; Hess, O.; Beermann, W. Phase specific function structure in spermatocyte nuclei of Drosophila melanogaster and their dependence of Y chromosomes. Chromosoma 1961, 12, 676–716. [Google Scholar] [CrossRef] [PubMed]
- Hardy, R.W.; Lindsley, D.L.; Livak, K.J.; Lewis, B.; Siversten, A.L.; Joslyn, G.L.; Edwards, J.; Bonaccorsi, S. Cytogenetic analysis of a segment of the Y chromosome of Drosophila melanogaster. Genetics 1984, 107, 591–610. [Google Scholar] [PubMed]
- Schmidt, A.; Palumbo, G.; Bozzetti, M.P.; Tritto, P.; Pimpinelli, S.; Schafer, U. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 1999, 151, 749–760. [Google Scholar] [PubMed]
- Tritto, P.; Specchia, V.; Fanti, L.; Berloco, M.; D’Alessandro, R.; Pimpinelli, S.; Palumbo, G.; Bozzetti, M.P. Structure, regulation and evolution of the crystal-Stellate system of Drosophila. Genetica 2003, 117, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Bozzetti, M.P.; Massari, S.; Finelli, P.; Meggio, F.; Pinna, L.A.; Boldyreff, B.; Issinger, O.G.; Palumbo, G.; Ciriaco, C.; Bonaccorsi, S.; et al. The Ste locus, a component of the parasitic cry-Ste system of Drosophila melanogaster, encodes a protein that forms crystals in primary spermatocytes and mimics properties of the beta subunit of casein kinase 2. Proc. Natl. Acad. Sci. USA 1995, 92, 6067–6071. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’Alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 2010, 463, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef]
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The small RNA profile during Drosophila melanogaster development. Dev. Cell 2003, 5, 337–350. [Google Scholar] [CrossRef]
- Stapleton, W.; Das, S.; McKee, B.D. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma 2001, 110, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Bozzetti, M.P.; Specchia, V.; Cattenoz, P.B.; Laneve, P.; Geusa, A.; Sahin, H.B.; Di Tommaso, S.; Friscini, A.; Massari, S.; Diebold, C.; et al. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J. Cell Sci. 2015, 128, 2070–2084. [Google Scholar] [CrossRef] [PubMed]
- Ishizu, H.; Siomi, H.; Siomi, M.C. Biology of PIWI-interacting RNAs: New insights into biogenesis and function inside and outside of germlines. Genes Dev. 2012, 26, 2361–2373. [Google Scholar] [CrossRef] [PubMed]
- Siomi, M.C.; Miyoshi, T.; Siomi, H. piRNA-mediated silencing in Drosophila germlines. Semin. Cell Dev. Biol. 2010, 21, 754–759. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Bozzetti, M.P. Different aubergine alleles confirm the specificity of different RNAi pathways in Drosophila melanogaster. Fly 2009, 3, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Kim, N.; de Planell-Saguer, M.; Khandros, E.; Chiorean, S.; Klein, P.S.; Rigoutsos, I.; Jongens, T.A.; Mourelatos, Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009, 11, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Chen, D.; Duan, R.; Xia, L.; Wang, J.; Qurashi, A.; Jin, P. Argonaute 1 regulates the fate of germline stem cells in Drosophila. Development 2007, 134, 4265–4272. [Google Scholar] [CrossRef] [PubMed]
- Mugat, B.; Akkouche, A.; Serrano, V.; Armenise, C.; Li, B.; Brun, C.; Fulga, T.A.; van Vactor, D.; Pelisson, A.; Chambeyron, S. MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells. PLoS Genet. 2015, 11, e1005194. [Google Scholar] [CrossRef] [PubMed]
- Cook, H.A.; Koppetsch, B.S.; Wu, J.; Theurkauf, W.E. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell 2004, 116, 817–829. [Google Scholar] [CrossRef]
- Handler, D.; Olivieri, D.; Novatchkova, M.; Gruber, F.S.; Meixner, K.; Mechtler, K.; Stark, A.; Sachidanandam, R.; Brennecke, J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011, 30, 3977–3993. [Google Scholar] [CrossRef] [PubMed]
- Zamparini, A.L.; Davis, M.Y.; Malone, C.D.; Vieira, E.; Zavadil, J.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 2011, 138, 4039–4050. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, D.; Senti, K.A.; Subramanian, S.; Sachidanandam, R.; Brennecke, J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 2012, 47, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Arkov, A.L.; Wang, J.Y.; Ramos, A.; Lehmann, R. The role of Tudor domains in germline development and polar granule architecture. Development 2006, 133, 4053–4062. [Google Scholar] [CrossRef] [PubMed]
- Nishida, K.M.; Okada, T.N.; Kawamura, T.; Mituyama, T.; Kawamura, Y.; Inagaki, S.; Huang, H.; Chen, D.; Kodama, T.; Siomi, H.; Siomi, M.C. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009, 28, 3820–3831. [Google Scholar] [CrossRef] [PubMed]
- Anne, J. Targeting and anchoring Tudor in the pole plasm of the Drosophila oocyte. PLoS ONE 2010, 5, e14362. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Anand, A.; Chakrabarti, A.; Kai, T. The Tudor domain protein Tapas, a homolog of the vertebrate Tdrd7, functions in piRNA pathway to regulate retrotransposons in germline of Drosophila melanogaster. BMC Biol. 2014, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Kai, T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2012, 31, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Qi, H.; Wang, J.; Lin, H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 2011, 138, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Malone, C.D.; Navarro, C.; Newbold, S.P.; Hayes, P.S.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 2011, 21, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Pek, J.W.; Lim, A.K.; Kai, T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev. Cell 2009, 17, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Anne, J.; Mechler, B.M. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development 2005, 132, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Anne, J.; Ollo, R.; Ephrussi, A.; Mechler, B.M. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 2007, 134, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Tomari, Y.; Du, T.; Zamore, P.D. Sorting of Drosophila small silencing RNAs. Cell 2007, 130, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Preall, J.B.; Czech, B.; Guzzardo, P.M.; Muerdter, F.; Hannon, G.J. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 2012, 18, 1446–1457. [Google Scholar] [CrossRef] [PubMed]
- Nagao, A.; Mituyama, T.; Huang, H.; Chen, D.; Siomi, M.C.; Siomi, H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 2010, 16, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.N.; Chao, A.; Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 2000, 127, 503–514. [Google Scholar] [PubMed]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; de Sapio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012, 149, 693–707. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.A.; Paquola, A.C.; Muotri, A.R. LINE-1 retrotransposition in the nervous system. Annu. Rev. Cell Dev. Biol. 2012, 28, 555–573. [Google Scholar] [CrossRef] [PubMed]
- Reilly, M.T.; Faulkner, G.J.; Dubnau, J.; Ponomarev, I.; Gage, F.H. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013, 33, 17577–17586. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P. The DEAD-box helicase Vasa: Evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim. Biophys. Acta 2013, 1829, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Kibanov, M.V.; Egorova, K.S.; Ryazansky, S.S.; Sokolova, O.A.; Kotov, A.A.; Olenkina, O.M.; Stolyarenko, A.D.; Gvozdev, V.A.; Olenina, L.V. A novel organelle, the piNG-body, in the nuage of Drosophila male germ cells is associated with piRNA-mediated gene silencing. Mol. Biol. Cell 2011, 22, 3410–3419. [Google Scholar] [CrossRef] [PubMed]
- Okray, Z.; de Esch, C.E.; Van Esch, H.; Devriendt, K.; Claeys, A.; Yan, J.; Verbeeck, J.; Froyen, G.; Willemsen, R.; de Vrij, F.M.; et al. A novel fragile X syndrome mutation reveals a conserved role for the carboxy-terminus in FMRP localization and function. EMBO Mol. Med. 2015, 7, 423–437. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Schauder, C.; Naryshkina, T.; Minakhina, S.; Steward, R. Zfrp8 forms a complex with fragile-X mental retardation protein and regulates its localization and function. Dev. Biol. 2016, 410, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Kirino, Y.; Vourekas, A.; Sayed, N.; de Lima Alves, F.; Thomson, T.; Lasko, P.; Rappsilber, J.; Jongens, T.A.; Mourelatos, Z. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 2010, 16, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Lu, F.; Li, P.; Liu, W.; Zhao, L.; Wang, Q.; Cao, X.; Zhang, L.; Zhang, Y.Q. Drosophila Homolog of FMRP Maintains Genome Integrity by Interacting with Piwi. J. Genet. Genom. 2016, 43, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Megosh, H.B.; Cox, D.N.; Campbell, C.; Lin, H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006, 16, 1884–1894. [Google Scholar] [CrossRef] [PubMed]
- Fanti, L.; Pimpinelli, S. HP1: A functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008, 18, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, S.; Xia, L.; Wang, J.; Wen, S.; Jin, P.; Chen, D. The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009, 5, e1000444. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Alisch, R.S.; Warren, S.T. RNA and microRNAs in fragile X mental retardation. Nat. Cell Biol. 2004, 6, 1048–1053. [Google Scholar] [CrossRef] [PubMed]
- Menon, L.; Mader, S.A.; Mihailescu, M.R. Fragile X mental retardation protein interactions with the microtubule associated protein 1B RNA. RNA 2008, 14, 1644–1655. [Google Scholar] [CrossRef] [PubMed]
- Pillai, R.S. MicroRNA function: Multiple mechanisms for a tiny RNA? RNA 2005, 11, 1753–1761. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Izaurralde, E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.; Jan, L.Y.; Jan, Y.N. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development 1990, 109, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P.F.; Ashburner, M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990, 4, 905–921. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Diehl-Jones, W.; Lasko, P. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 1994, 120, 1201–1211. [Google Scholar] [PubMed]
- Findley, S.D.; Tamanaha, M.; Clegg, N.J.; Ruohola-Baker, H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 2003, 130, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Khurana, J.S.; Theurkauf, W. piRNAs, transposon silencing, and Drosophila germline development. J. Cell Biol. 2010, 191, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, G.; Calhoun, G.; Schedl, P. The drosophila fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics 2006, 174, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, M.; Lasko, P. In vivo mapping of the functional regions of the DEAD-box helicase Vasa. Biol. Open 2015, 4, 450–462. [Google Scholar] [CrossRef] [PubMed]
- Verrotti, A.C.; Wharton, R.P. Nanos interacts with cup in the female germline of Drosophila. Development 2000, 127, 5225–5232. [Google Scholar] [PubMed]
- Zappavigna, V.; Piccioni, F.; Villaescusa, J.C.; Verrotti, A.C. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc. Natl. Acad. Sci. USA 2004, 101, 14800–14805. [Google Scholar] [CrossRef] [PubMed]
- Pisa, V.; Cozzolino, M.; Gargiulo, S.; Ottone, C.; Piccioni, F.; Monti, M.; Gigliotti, S.; Talamo, F.; Graziani, F.; Pucci, P.; Verrotti, A.C. The molecular chaperone Hsp90 is a component of the cap-binding complex and interacts with the translational repressor Cup during Drosophila oogenesis. Gene 2009, 432, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Cziko, A.M.; McCann, C.T.; Howlett, I.C.; Barbee, S.A.; Duncan, R.P.; Luedemann, R.; Zarnescu, D.; Zinsmaier, K.E.; Parker, R.R.; Ramaswami, M. Genetic modifiers of dFMR1 encode RNA granule components in Drosophila. Genetics 2009, 182, 1051–1060. [Google Scholar] [CrossRef] [PubMed]
- Ottone, C.; Gigliotti, S.; Giangrande, A.; Graziani, F.; Verrotti di Pianella, A. The translational repressor Cup is required for germ cell development in Drosophila. J. Cell Sci. 2012, 125 Pt 13, 3114–3123. [Google Scholar] [CrossRef] [PubMed]
- Sawarkar, R.; Sievers, C.; Paro, R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell 2012, 149, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Karam, J.A.; Parikh, R.Y.; Nayak, D.; Rosenkranz, D.; Gangaraju, V.K. Co-chaperone Hsp70/Hsp90 organizing protein (Hop) is Required for Transposon Silencing and piRNA Biogenesis. J. Biol. Chem. 2017, 292, 6039–6046. [Google Scholar] [CrossRef] [PubMed]
- Piacentini, L.; Fanti, L.; Specchia, V.; Bozzetti, M.P.; Berloco, M.; Palumbo, G.; Pimpinelli, S. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma 2014, 123, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Thomson, T.; Liu, N.; Arkov, A.; Lehmann, R.; Lasko, P. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008, 125, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, J.; Barbee, S.A.; Ramaswami, M. P-body components, microRNA regulation, and synaptic plasticity. Sci. World J. 2007, 7, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Barbee, S.A.; Estes, P.S.; Cziko, A.M.; Hillebrand, J.; Luedeman, R.A.; Coller, J.M.; Johnson, N.; Howlett, I.C.; Geng, C.; Ueda, R.; et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006, 52, 997–1009. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Izaurralde, E. P bodies: At the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 2007, 8, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.C.; Jensen, K.B.; Jin, P.; Brown, V.; Warren, S.T.; Darnell, R.B. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 2001, 107, 489–499. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Xie, H.; Zhou, W.; Wu, Y.; Qin, J.; Guo, J.; Gu, Q.; Zhang, X.; Ji, T.; et al. Fragile X syndrome screening in Chinese children with unknown intellectual developmental disorder. BMC Pediatr. 2015, 15, 77. [Google Scholar] [CrossRef] [PubMed]
- Bardoni, B.; Willemsen, R.; Weiler, I.J.; Schenck, A.; Severijnen, L.A.; Hindelang, C.; Lalli, E.; Mandel, J.L. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp. Cell Res. 2003, 289, 95–107. [Google Scholar] [CrossRef]
- Cabart, P.; Chew, H.K.; Murphy, S. BRCA1 cooperates with NUFIP and P-TEFb to activate transcription by RNA polymerase II. Oncogene 2004, 23, 5316–5329. [Google Scholar] [CrossRef] [PubMed]
- Rothe, B.; Saliou, J.M.; Quinternet, M.; Back, R.; Tiotiu, D.; Jacquemin, C.; Loegler, C.; Schlotter, F.; Pena, V.; Eckert, K.; et al. Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res. 2014, 42, 10731–10747. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Changela, N.; Steward, R. Zfrp8/PDCD2 is required in ovarian stem cells and interacts with the piRNA pathway machinery. Development 2014, 141, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Minakhina, S.; Steward, R. Hematopoietic stem cells in Drosophila. Development 2010, 137, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Yin, H.; Weiner, M.M.; Wang, J.; Huang, X.A.; Lin, H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 2011, 43, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 2013, 25, 190–194. [Google Scholar] [CrossRef] [PubMed]
| Mutants | Protein Domains | Protein Localization | Testes | Ovaries | Protein Function | Transposons | Stellate-Made Crystals | References |
|---|---|---|---|---|---|---|---|---|
| Argonaute Proteins | ||||||||
| aubergine | PAZ, MID, PIWI | germline (C-nuage); germ granules | + * | + | ping-pong pathway, primary pathway | germinal | + | [35,38,78,83,88,108] |
| ago3 | PAZ, MID, PIWI | germline (C-nuage); germ granules | + * | + | ping-pong pathway | germinal | + | [35,70,71] |
| piwi | PAZ, MID, PIWI | soma + germline (nuclear) | + | + | primary pathway | germinal + somatic | - | [83,84,109] |
| ago1 | PAZ, PIWI | soma, germline (C-nuage) | + * | + | piRNA pathway, miRNA pathway | somatic | + | [105,111] |
| Helicases | ||||||||
| armitage | RNA helicase (SDE3) | soma(C-Yb bodies) + germline (C) | + | + | primary pathway | germinal + somatic | + | [70,71,112,113,114] |
| vasa | DEAD RNA helicase | germline (Amplifier Complex-C-nuage); germ granules | + * | + | primary + ping-pong pathway | germinal + somatic | ++ (RNA) | [84,86] |
| UAP56 | DEAD RNA helicase | germline (N) + N/C boundary | ND | + | primary + ping-pong pathway | germinal + somatic | ND | [73,75] |
| Tudor Proteins | ||||||||
| Yb | DEAD RNA helicase + Tudor domain | soma (C-Yb bodies) | ND | + | primary pathway (single strand clusters (flamenco)) | somatic | - | [32,70,71,113,114] |
| spindle-E | DEAD RNA helicase + Tudor domain | germline (C-nuage) | + * | + | primary pathway | germinal | + | [61,84,104] |
| krimper | Tudor domain | germline (C-nuage) + soma (krimper body) | + | + | germinal | ++++ | [61,115] | |
| tudor | Tudor domain | germline (C-nuage) soma; germ granules | + * | + | germinal | - | [116,117,118] | |
| tapas | Tudor domain + Lotus | germline (C-nuage) | + | + | ping-pong?, primary? | germinal | ++++ (RNA) | [113,119] |
| tejas | Tudor domain + Lotus domain | germline (C-nuage) | + | + | ping-pong?, primary? | germinal | ++++ | [84] |
| zucchini | Tudor domain + nuclease | soma(C) + germline (C-nuage) mitocondrial | + | + | primary pathway | germinal + somatic | + | [65,66,69,71,81] |
| squash | Tudor domain + nuclease | germline (C-nuage); ND in the soma | + * | + | germinal + somatic | + | [69,81] | |
| vreteno | Tudor domain + RRM domai | soma (C) + germline (C-nuage) | + | + | primary pathway | germinal (seen only in ovaries) + somatic | ND | [113,114] |
| qin/kumo | Tudor domain + RING domain | soma (N) + germline (Amplifier Complex-C-nuage) | + | + | primary pathway (dual strand clusters) | germline + somatic | + | [75,85,120] |
| dFmr1 | Tudor domain, KH, RGG, NLS, NES | soma + germline (N + C) | + | + | germinal + somatic | + | [105] | |
| papi | Tudor domain + KH domain | soma (N) + germline (C-nuage) | ND | + | primary + ping-pong pathway | germinal + somatic | ND | [121] |
| eggless (dSETDB1) | Tudor domain + SET domain | germline (N) + soma (N) | ND | + | H3K9 methyl transferase (TE regulation) | germinal + somatic | ND | [122] |
| Non Tudor Proteins | ||||||||
| rhino | chromo/shadow domain | germline + soma (N) | ND | + | primary + ping-pong pathway (dual strand clusters) | germinal | - | [57,76,82] |
| cutoff | DXO/Dom3Z family | germline (C-nuage) + N | ND | + | primary + ping-pong pathway (dual strand clusters) | germinal | - | [57,76] |
| maelstrom | Maelstrom domain (DNA/RNA binding); HMG protein | germline + soma: shuttle nucleus-cytoplasm (nuage) | + | + | primary + ping-pong pathway | germinal + somatic | ND | [123] |
| capsuleen | PRMT | germline (C-nuage) | + | + | arginine methyltransferase | germline | + | [124,125] |
| shutdown | FKBP6 family (co-chaperone) | soma (YB-bodies) + germline (N + C) | + | + | primary pathway + ping-pong | germinal + somatic | + | [115,127] |
| hsp83 | heat shock protein | soma + germline (N + C) | + | + | primary pathway + ping-pong | germinal + somatic | + | [70,78,99,101] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Specchia, V.; D’Attis, S.; Puricella, A.; Bozzetti, M.P. dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster. Int. J. Mol. Sci. 2017, 18, 1066. https://doi.org/10.3390/ijms18051066
Specchia V, D’Attis S, Puricella A, Bozzetti MP. dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster. International Journal of Molecular Sciences. 2017; 18(5):1066. https://doi.org/10.3390/ijms18051066
Chicago/Turabian StyleSpecchia, Valeria, Simona D’Attis, Antonietta Puricella, and Maria Pia Bozzetti. 2017. "dFmr1 Plays Roles in Small RNA Pathways of Drosophila melanogaster" International Journal of Molecular Sciences 18, no. 5: 1066. https://doi.org/10.3390/ijms18051066