Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus
Abstract
:1. Introduction
2. Results
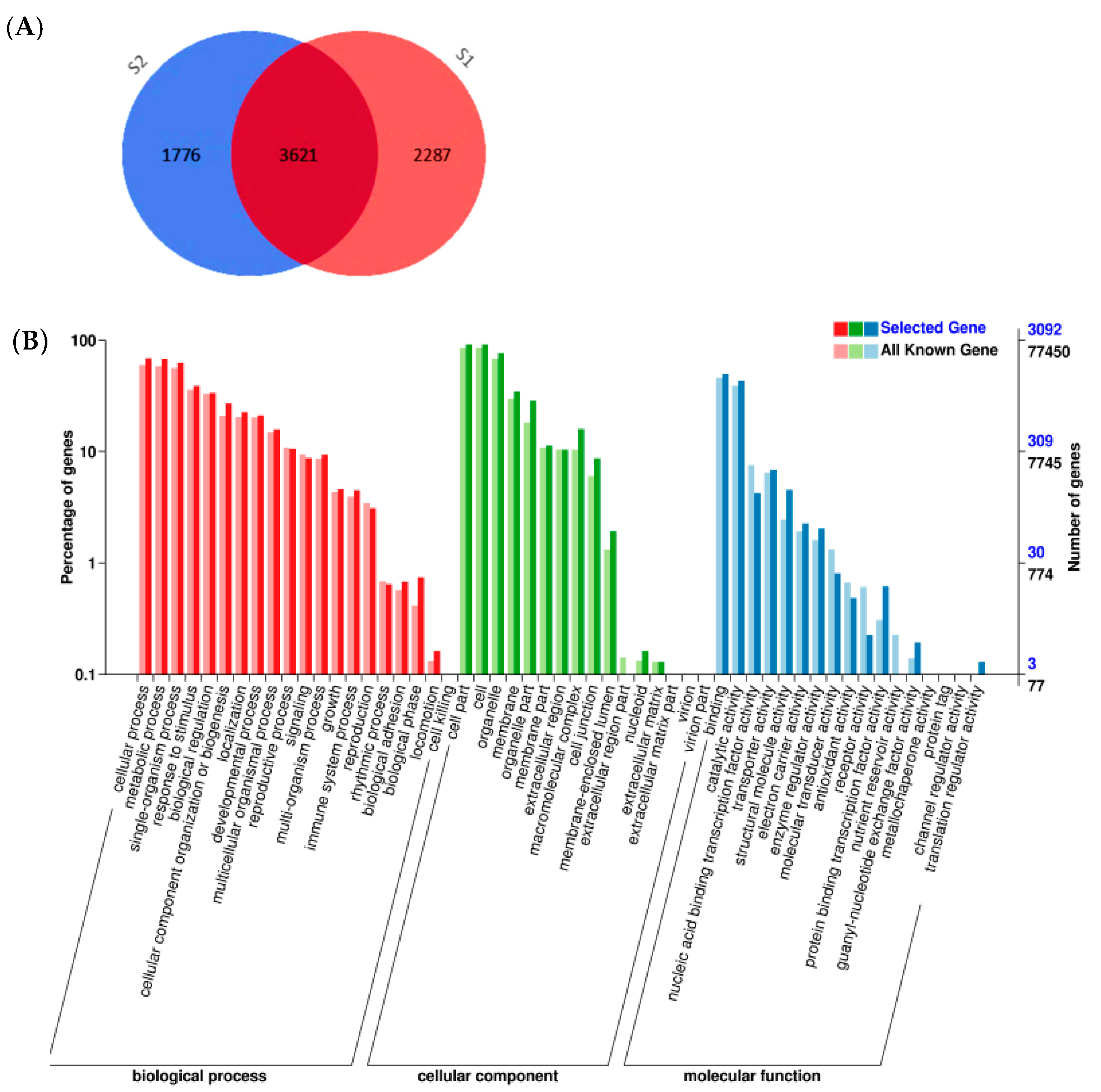
2.1. Differentially Expressed Gene Analysis by RNA-Seq
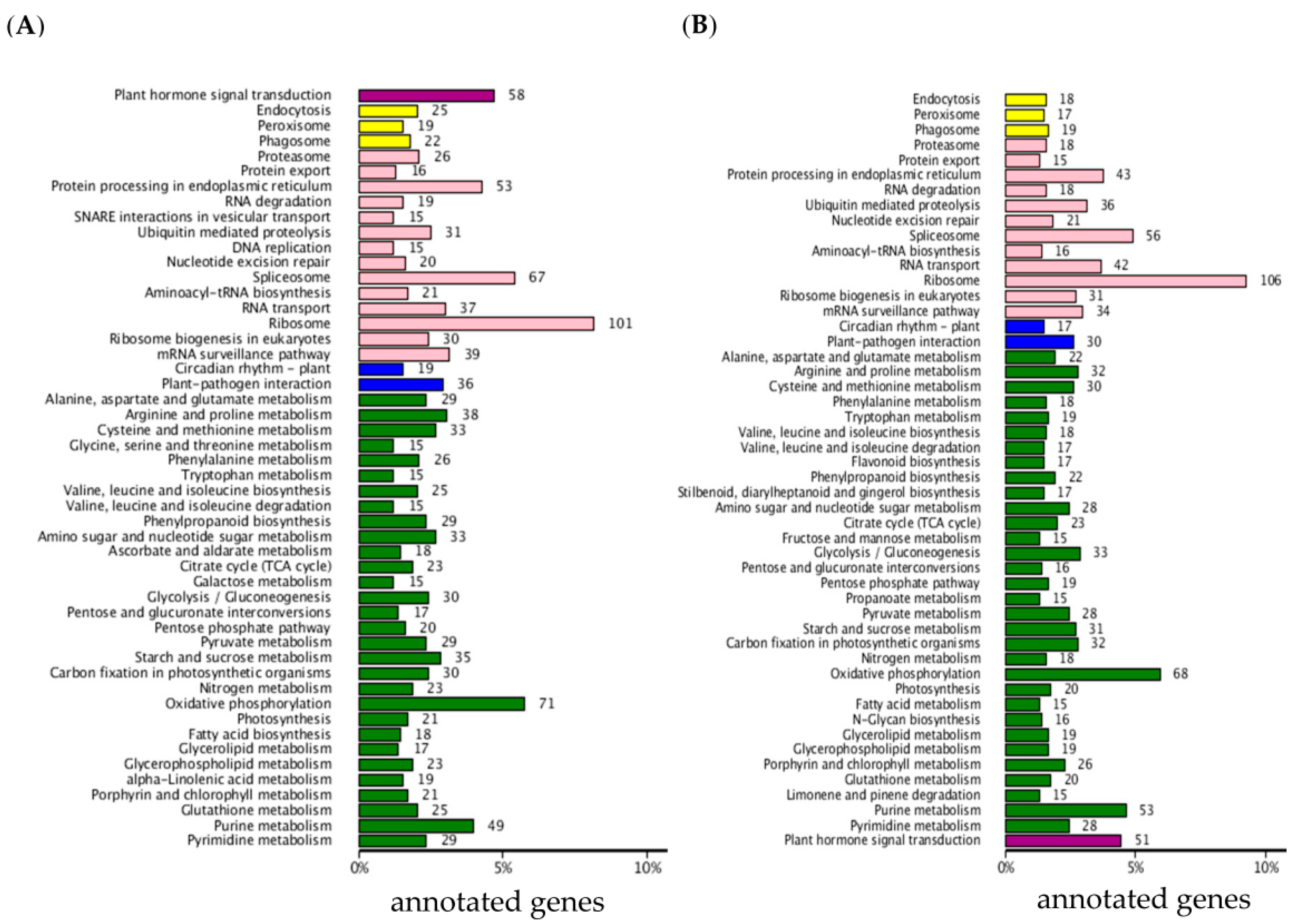
2.2. Functional Classification by Gene Ontology and Metabolic Pathway Analysis
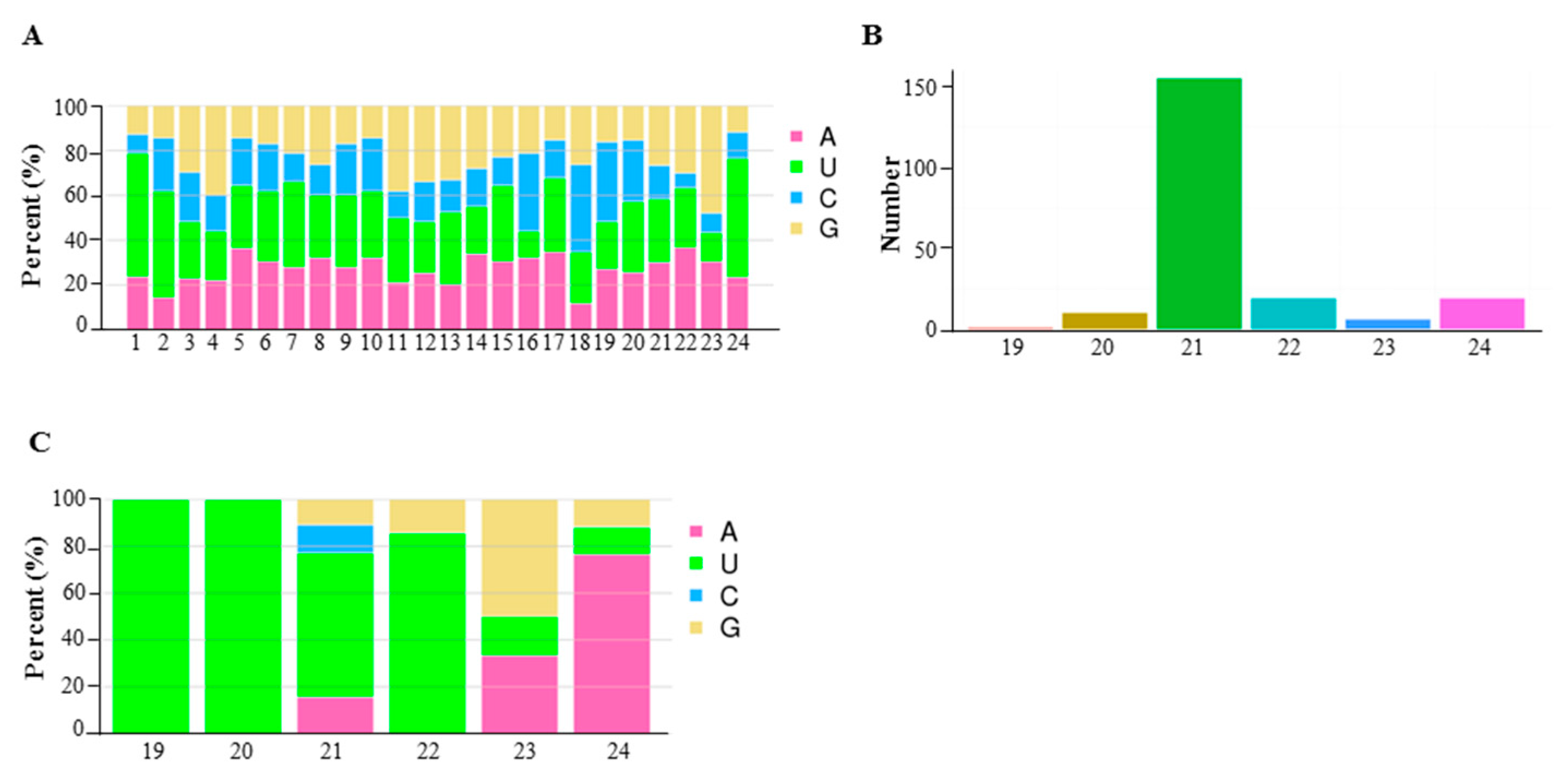
2.3. Differentially Expressed miRNA in 6098B and Purler
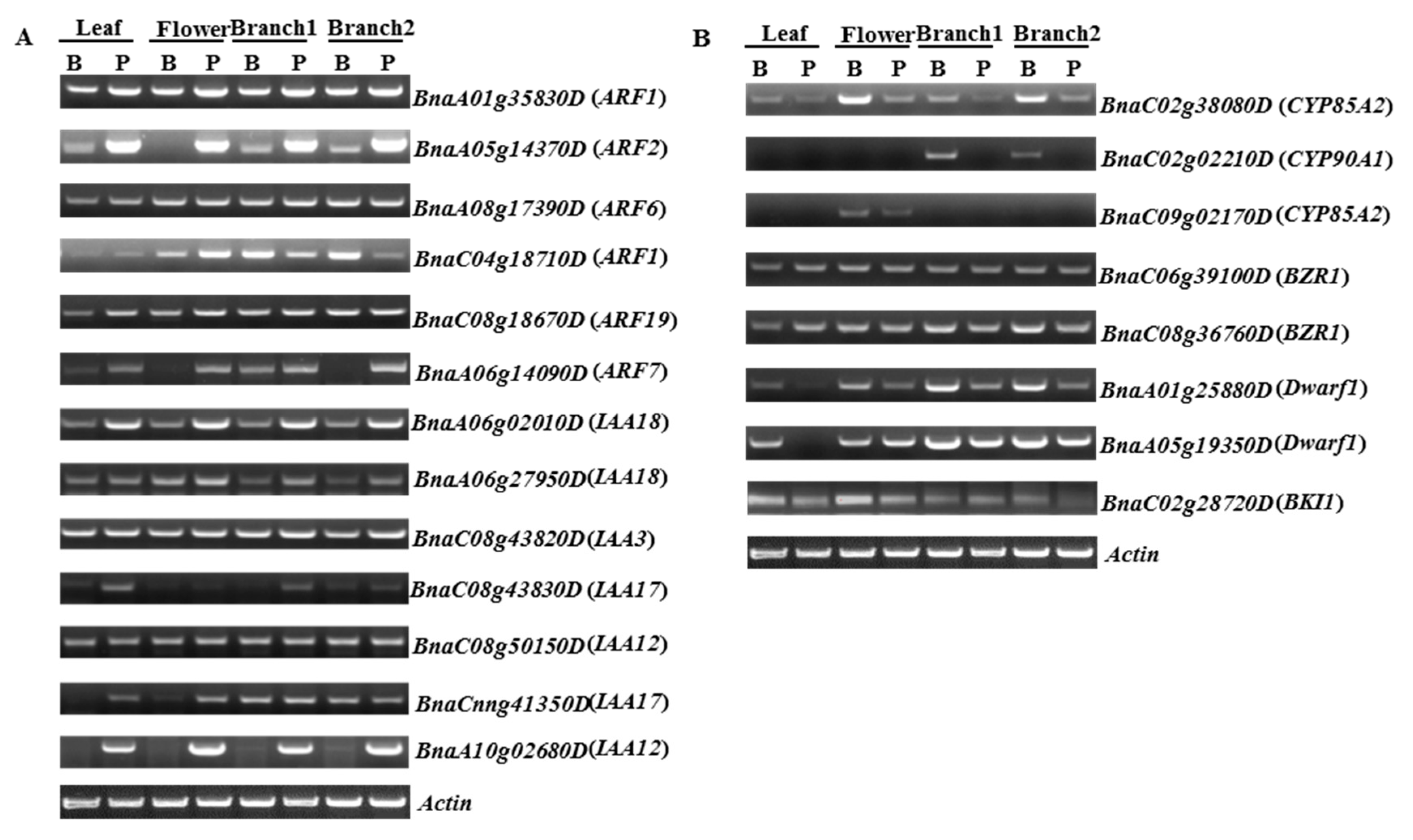
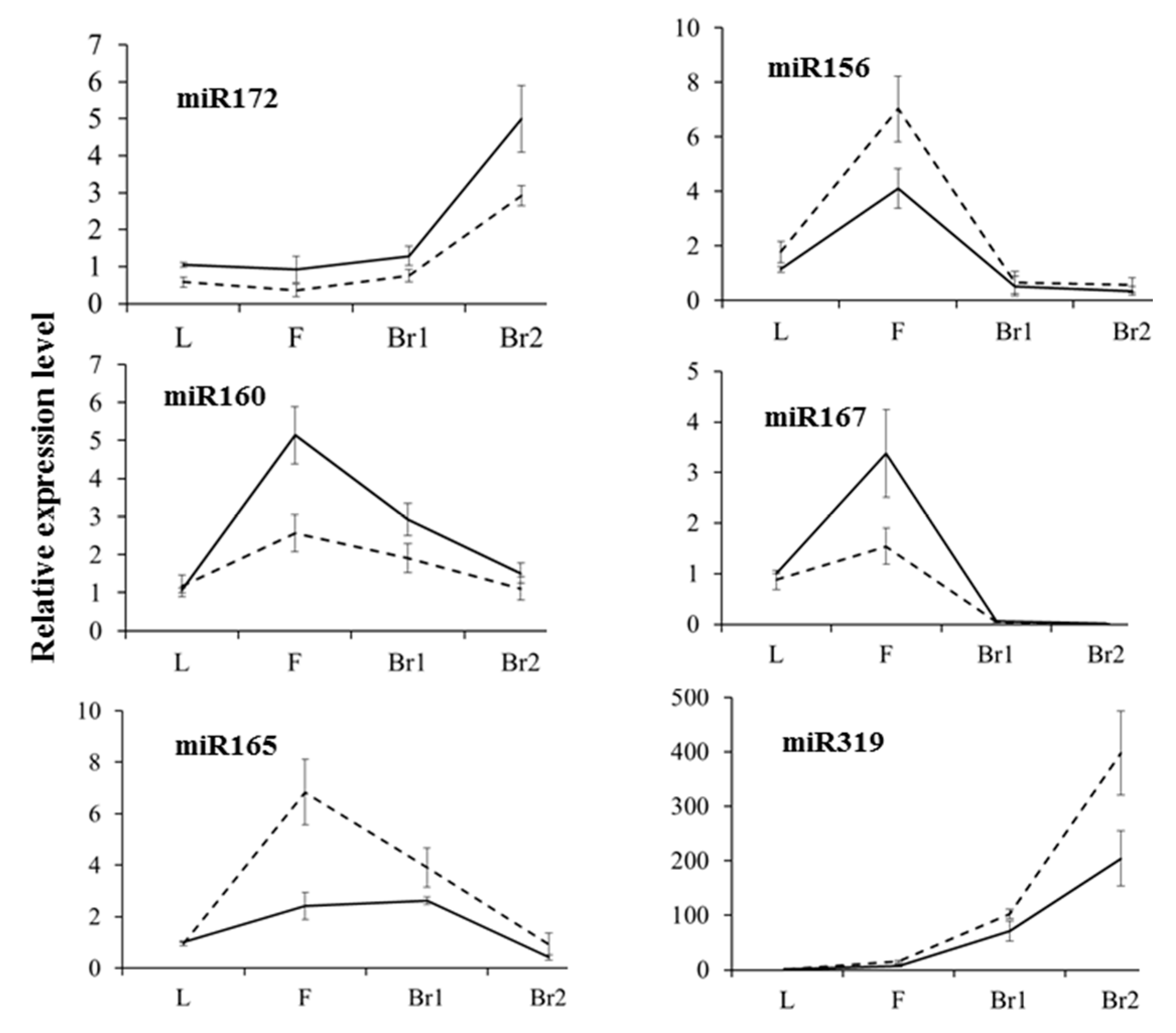
2.4. Validation of Differential Gene and miRNA Expression by semi-RT-PCR and qRT-PCR
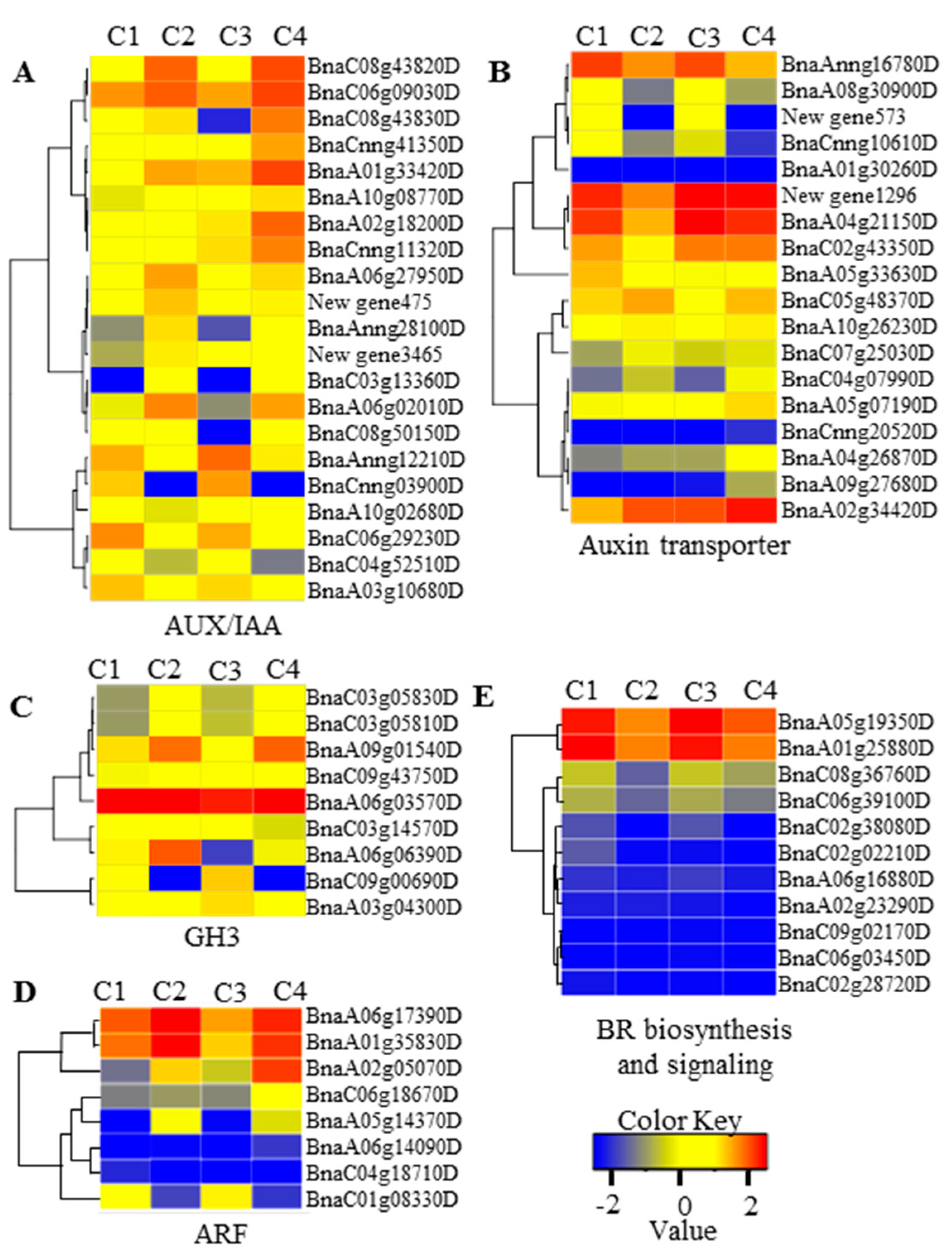
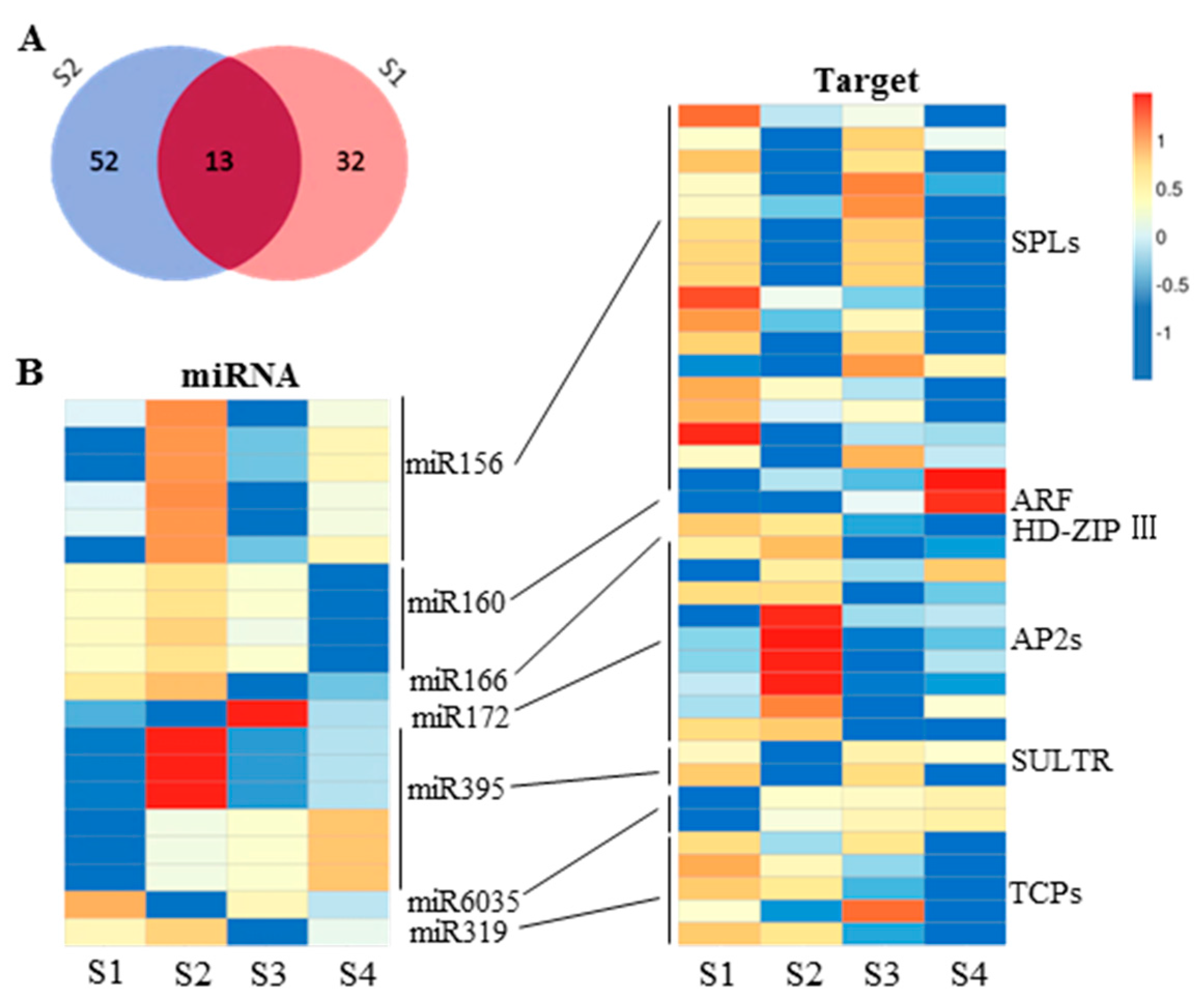
2.5. miRNA and Gene Network Analysis in 6098B and Purler
3. Discussion
3.1. Branch Angle Regulation in B. napus and Other Species
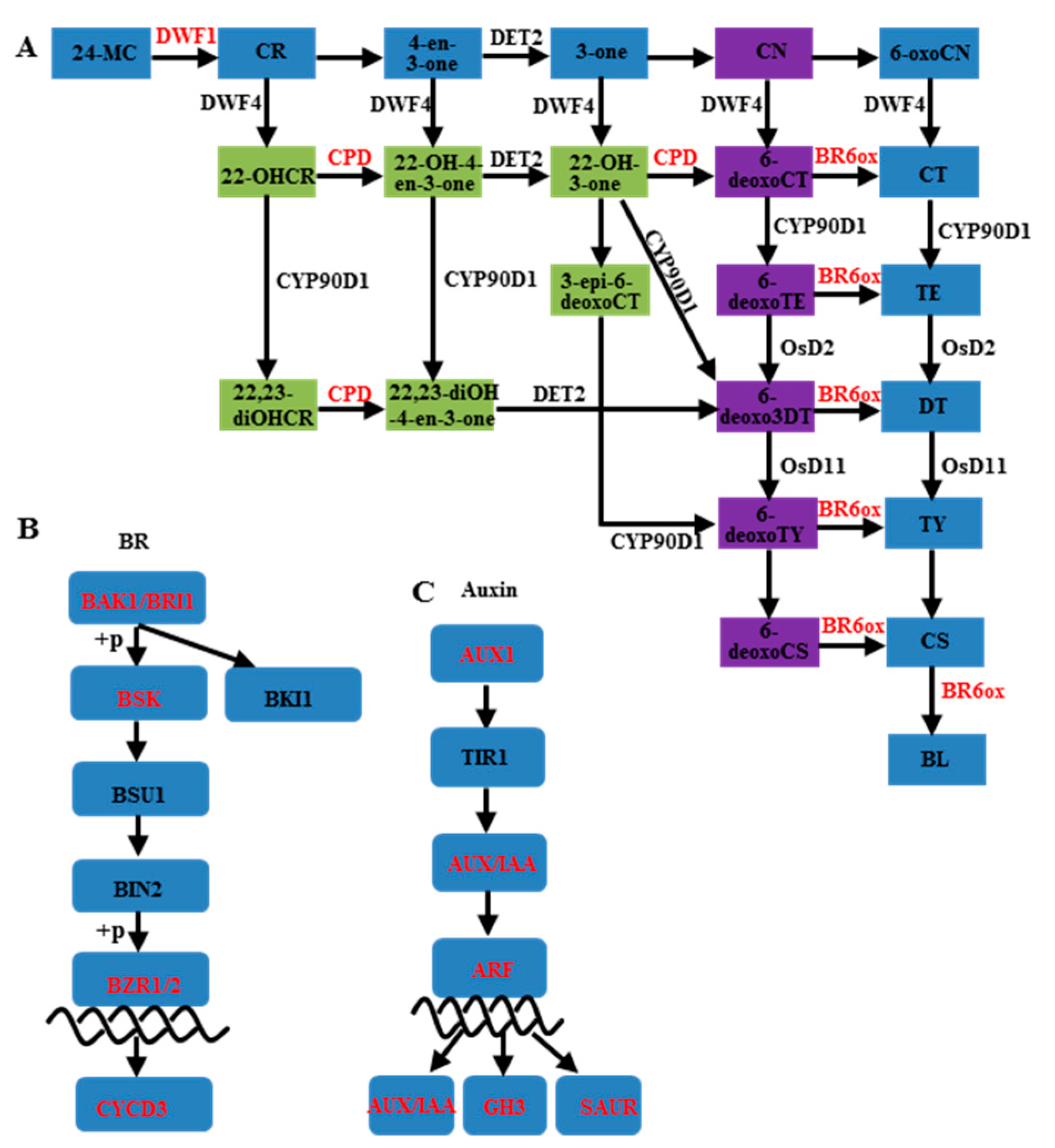
3.2. Auxin and Brassinosteroids in Branch Angle and Leaf Inclination Regulation
3.3. Involvement of miRNA in Plant Architecture Establishment
4. Materials and Methods
4.1. Plant Materials and RNA Preparation
4.2. Transcriptome Sequencing and Gene Expression Analysis
4.3. Identification of Known and Novel miRNAs
4.4. Real-Time RT-PCR
Supplementary Materials
Acknowledgment
Author Contributions
Conflicts of Interest
Abbreviations
| BR | Brassinosteroid |
| TCP | TEOSINTE-BRANCHED/CYCLOIDEA/PCF |
| ARF | Auxin response factor |
| DEG | Differentially expressed gene |
| NAC | NAM, ATAF, and CUC |
| IAA | Indole-3-acetic acid |
| SULT | Sulfate Transporter |
References
- Mansfield, B.D.; Mumm, R.H. Survey of plant density tolerance in US maize germplasm. Crop Sci. 2014, 54, 157–173. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Brutnell, T.P. The molecular analysis of the shade avoidance syndrome in the grasses has begun. J. Exp. Bot. 2007, 58, 3079–3089. [Google Scholar] [CrossRef] [PubMed]
- Nik, M.M.; Babaeian, M.; Tavassoli, A.; Asgharzade, A. Effect of plant density on yield and yield components of corn hybrids (Zea mays). Sci. Res. Essays 2011, 6, 4821–4825. [Google Scholar]
- Duvick, D.N. Genetic progress in yield of United States maize (Zea mays L.). Maydica 2005, 50, 193–202. [Google Scholar]
- Reinhardt, D.; Kuhlemeier, C. Plant architecture. EMBO Rep. 2002, 3, 846–851. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Branching in rice. Curr. Opin. Plant Biol. 2011, 14, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, J. Genes controlling plant architecture. Curr. Opin. Biotechnol. 2006, 17, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Roychoudhry, S.; Kepinski, S. Shoot and root branch growth angle control—The wonderfulness of lateralness. Curr. Opin. Plant Biol. 2015, 23, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, Y.; Qian, Q.; Fu, Z.; Wang, M.; Zeng, D.; Li, B.; Wang, X.; Li, J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007, 17, 402–410. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, X.; Liu, F.; Sun, X.; Li, C.; Zhu, Z.; Fu, Y.; Cai, H.; Wang, X.; Xie, D.; et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 2008, 40, 1360–1364. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tang, D.; Li, M.; Wang, K.; Cheng, Z. Loose plant architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol. 2013, 161, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Ku, L.; Wei, X.; Zhang, S.; Zhang, J.; Guo, S.; Chen, Y. Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 2011, 6, e20621. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Spalding, E.p.; Iino, M. AtLAZY1 is a signaling component required for gravitropism of the Arabidopsis thaliana inflorescence. Plant J. 2013, 74, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.; Meng, X.; Chu, J.; Sun, X.; Dong, G.; Yuan, Y.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef] [PubMed]
- Harmoko, R.; Yoo, J.Y.; Ko, K.S.; Ramasamy, N.K.; Hwang, B.Y.; Lee, E.J.; Kim, H.S.; Lee, K.J.; Oh, D.B.; Kim, D.Y.; et al. N-glycan containing a core α1,3-fucose residue is required for basipetal auxin transport and gravitropic response in rice (Oryza sativa). New Phytol. 2016, 212, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Fujioka, S.; Takatsuto, S.; Tsujimoto, M.; Kitano, H.; Oshida, S.; Asami, T.; Nakano, T. Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint ending in rice. Plant Cell Physiol. 2009, 50, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Fouracre, J.P.; Poethig, R.S. The role of small RNAs in vegetative shoot development. Curr. Opin. Plant Biol. 2016, 29, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutiérrez, R.A. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F., Jr.; Vazquez, F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011, 157, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Han, S.J.; Yoon, E.K.; Lee, W.S. Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 2006, 34, 1892–1899. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.J.; Wartell, R.M.; Cairney, J.; Pullman, G.S. Evidence for stage-specific modulation of specific microRNAs (miRNAs) and miRNA processing components in zygotic embryo and female gametophyte of loblolly pine (Pinus taeda). New Phytol. 2008, 179, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genom. 2011, 12, 178. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Chen, Z.H.; Chen, G.X.; Xiong, L.Z.; Wu, C.Y. Study of overexpressing miRNA167a to regulate the architecture in Oryza sativa. Mol. Plant Breed. 2011, 9, 390–396. [Google Scholar]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.K.; Yang, J.H.; Lim, J.; Kim, S.H.; Kim, S.K.; Lee, W.S. Auxin regulation of the microRNA390-dependent transacting small interfering RNA pathway in Arabidopsis lateral root development. Nucleic Acids Res. 2010, 38, 1382–1391. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, W.; Mei, D.; Wang, H.; Fu, L.; Liu, D.; Li, Y.; Hu, Q. Characterizing Variation of Branch Angle and Genome-Wide Association Mapping in Rapeseed (Brassica napus L.). Front. Plant Sci. 2016, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, B.; Wang, X.; Hu, K.; Li, K.; Li, Z.; Li, S.; Yan, L.; Guan, C.; Zhang, J.; et al. Genome-Wide Association Study Dissecting the Genetic Architecture Underlying the Branch Angle Trait in Rapeseed (Brassica napus L.). Sci. Rep. 2016, 6, 33673. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, H.; Wang, W.; Liu, J.; Hao, M.; Mei, D.; Zhou, R.; Fu, L.; Hu, Q. Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-seq. Sci. Rep. 2016, 6, 38493. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Shen, E.; Zou, J.; Hubertus, B.F.; Chen, L.; Ye, C.; Dai, S.; Li, R.; Ni, M.; Jiang, X.; Qiu, J.; et al. Identification, evolution, and expression partitioning of miRNAs in allopolyploid Brassica napus. J. Exp. Bot. 2015, 66, 7241–7253. [Google Scholar] [CrossRef] [PubMed]
- Van der Honing, H.S.; Kieft, H.; Emons, A.M.; Ketelaar, T. Arabidopsis VILLIN2 and VILLIN3 are required for the generation of thick actin filament bundles and for directional organ growth. Plant Physiol. 2012, 158, 1426–1438. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Xie, Y.; Zhang, J.; Ren, Y.; Zhang, X.; Wang, J.; Guo, X.; Wu, F.; Sheng, P.; Wang, J.; et al. VLN2 Regulates Plant Architecture by Affecting Microfilament Dynamics and Polar Auxin Transport in Rice. Plant Cell 2015, 27, 2829–2845. [Google Scholar] [PubMed]
- Kuai, J.; Sun, Y.; Zuo, Q.; Huang, H.; Liao, Q.; Wu, C.; Lu, J.; Wu, J.; Zhou, G. The yield of mechanically harvested rapeseed (Brassica napus L.) can be increased by optimum plant density and row spacing. Sci. Rep. 2015, 5, 18835. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Li, J.; Zhang, W.; Yan, S.; Wang, R.; Zhao, J.; Li, Y.; Qi, Z.; Sun, Z.; Zhu, Z. The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance. Plant J. 2012, 72, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Cao, Y.; Huang, L.; Zhao, J.; Xu, C.; Li, X.; Wang, S. Activation of the indole-3-acetic acid-amido synthetase GH3-8 suppresses expansin expression and promotes salicylateand jasmonate-independent basal immunity in rice. Plant Cell 2008, 20, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Liu, H.; Li, Y.; Yu, H.; Li, X.; Xiao, J.; Wang, S. Manipulating broad-spectrum disease resistance by suppressing pathogen-induced auxin accumulation in rice. Plant Physiol. 2011, 155, 589–602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xiang, J.; Xue, H. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant 2013, 6, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Yamamuro, C.; Ihara, Y.; Wu, X.; Noguchi, T.; Fujioka, S.; Takatsuto, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 2000, 12, 1591–1606. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Ueguchi-Tanaka, M.; Matsuoka, M. Brassinosteroids and rice architecture. J. Pestic. Sci. 2004, 29, 184–187. [Google Scholar] [CrossRef]
- Bai, M.; Zhang, L.; Gampala, S.; Zhu, S.; Song, W.; Chong, K.; Wang, Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 13839–13844. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Yin, Y.; Fei, S. Brassinosteroid signaling network: Implications on yield and stress tolerance. Plant Cell Rep. 2013, 32, 1017–1030. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin, a master regulator in plant root development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rowe, J.; Lindsey, K. Hormonal crosstalk for root development: A combined experimental and modeling perspective. Front. Plant Sci. 2014, 5, 116. [Google Scholar] [CrossRef] [PubMed]
- Chaiwanon, J.; Wang, Z.Y. Spatiotemporal brassinosteroid signaling and antagonism with auxin pattern stem cell dynamics in Arabidopsis roots. Curr. Biol. 2015, 25, 1031–1042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front. Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Ou, X.; Tang, H.; Wang, R.; Wu, P.; Jia, Y.; Wei, X.; Xu, X.; Kang, S.H.; Kim, S.K.; et al. Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015, 208, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Bian, H.; Xie, Y.; Guo, F.; Han, N.; Ma, S.; Zeng, Z.; Wang, J.; Yang, Y.; Zhu, M. Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 2012, 196, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tan, L.; Fu, Y.; Liu, F.; Cai, H.; Xie, D.; Wu, F.; Wu, J.; Matsumoto, T.; Sun, C. Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 2013, 4, 2200. [Google Scholar] [CrossRef] [PubMed]
- Schenck, C.A.; Nadella, V.; Clay, S.L.; Lindner, J.; Abrams, Z.; Wyatt, S.E. A proteomics approach identifies novel proteins involved in gravitropic signal transduction. Am. J. Bot. 2013, 100, 194–202. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Yang, Z.; Yi, B.; Wen, J.; Shen, J.; Tu, J.; Ma, C.; Fu, T. Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus. BMC Genom. 2014, 15, 258. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jiang, L.; Wang, J.; Gu, P.; Chen, M. MTide: An integrated tool for the identification of miRNA target interaction in plants. Bioinformatics 2015, 31, 290–291. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. MicroRNA directed phasing during transacting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xie, K.; Xiong, L. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol. Genet. Genom. 2010, 284, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, M.; Fu, S.; Yang, W.; Qi, C.; Wang, X. Small RNA profiling in two Brassica napus cultivars identifies microRNAs with oil production- and development-correlated expression and new small RNA classes. Plant Physiol. 2012, 158, 813–823. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Wells, R.; Liu, J.; Wang, H.; Sang, S.; Tang, M.; Zhou, R.; et al. Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus. Int. J. Mol. Sci. 2017, 18, 887. https://doi.org/10.3390/ijms18050887
Cheng H, Hao M, Wang W, Mei D, Wells R, Liu J, Wang H, Sang S, Tang M, Zhou R, et al. Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus. International Journal of Molecular Sciences. 2017; 18(5):887. https://doi.org/10.3390/ijms18050887
Chicago/Turabian StyleCheng, Hongtao, Mengyu Hao, Wenxiang Wang, Desheng Mei, Rachel Wells, Jia Liu, Hui Wang, Shifei Sang, Min Tang, Rijin Zhou, and et al. 2017. "Integrative RNA- and miRNA-Profile Analysis Reveals a Likely Role of BR and Auxin Signaling in Branch Angle Regulation of B. napus" International Journal of Molecular Sciences 18, no. 5: 887. https://doi.org/10.3390/ijms18050887





