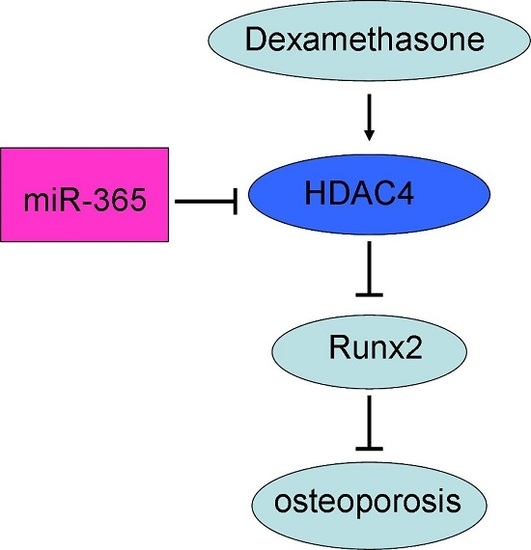
miR-365 Ameliorates Dexamethasone-Induced Suppression of Osteogenesis in MC3T3-E1 Cells by Targeting HDAC4
Abstract
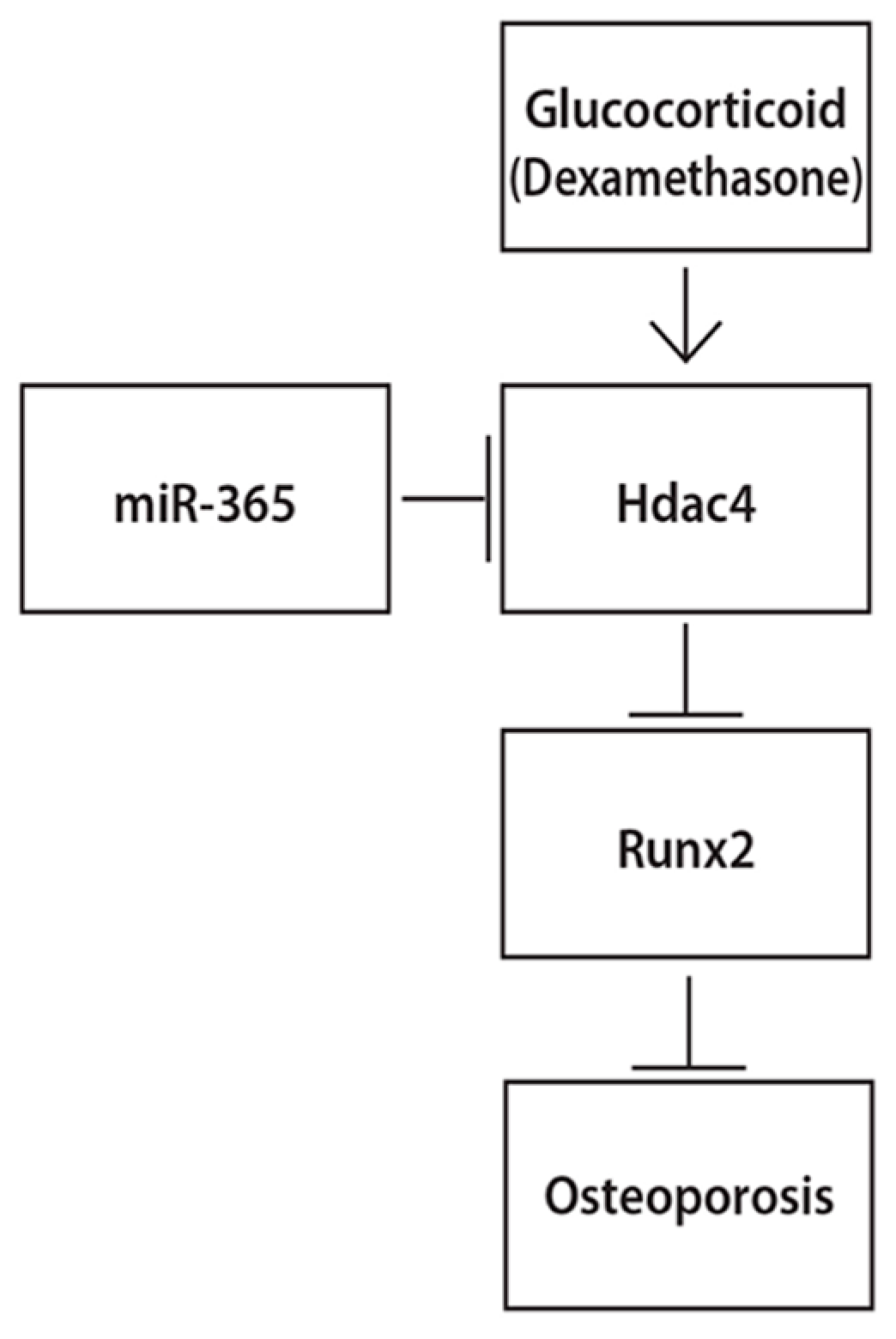
:1. Introduction
2. Results
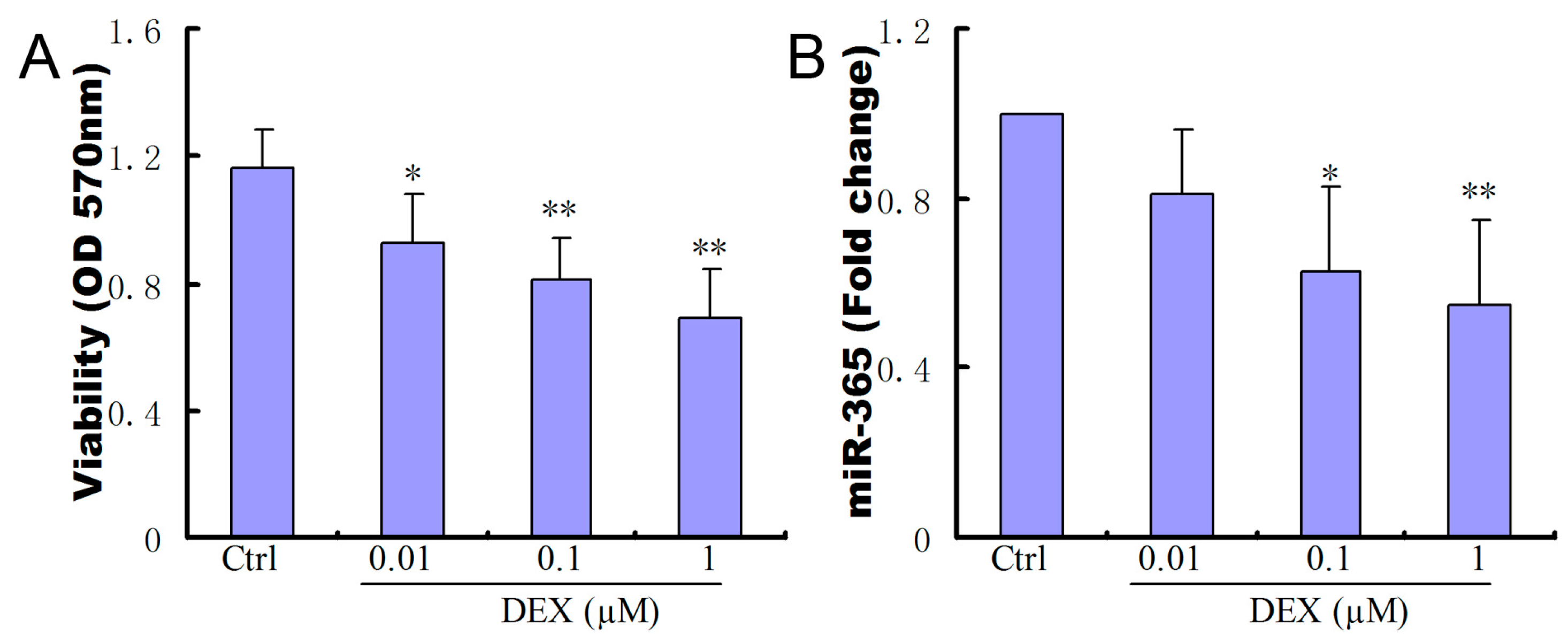
2.1. Dexamethasone (DEX) Inhibited Cell Viability and Decreased the Expression of miR-365 in MC3T3-E1 Cells
2.2. MiR-365 Over-Expression Ameliorated DEX-Induced Inhibition of Osteoblast Cell Viability and Alkaline Phosphatase (ALP) Activity
2.3. MiR-365 Over-Expression Attenuated the Suppressive Effect of DEX on Osteogenic Genes Expression in MC3T3-E1 Cells
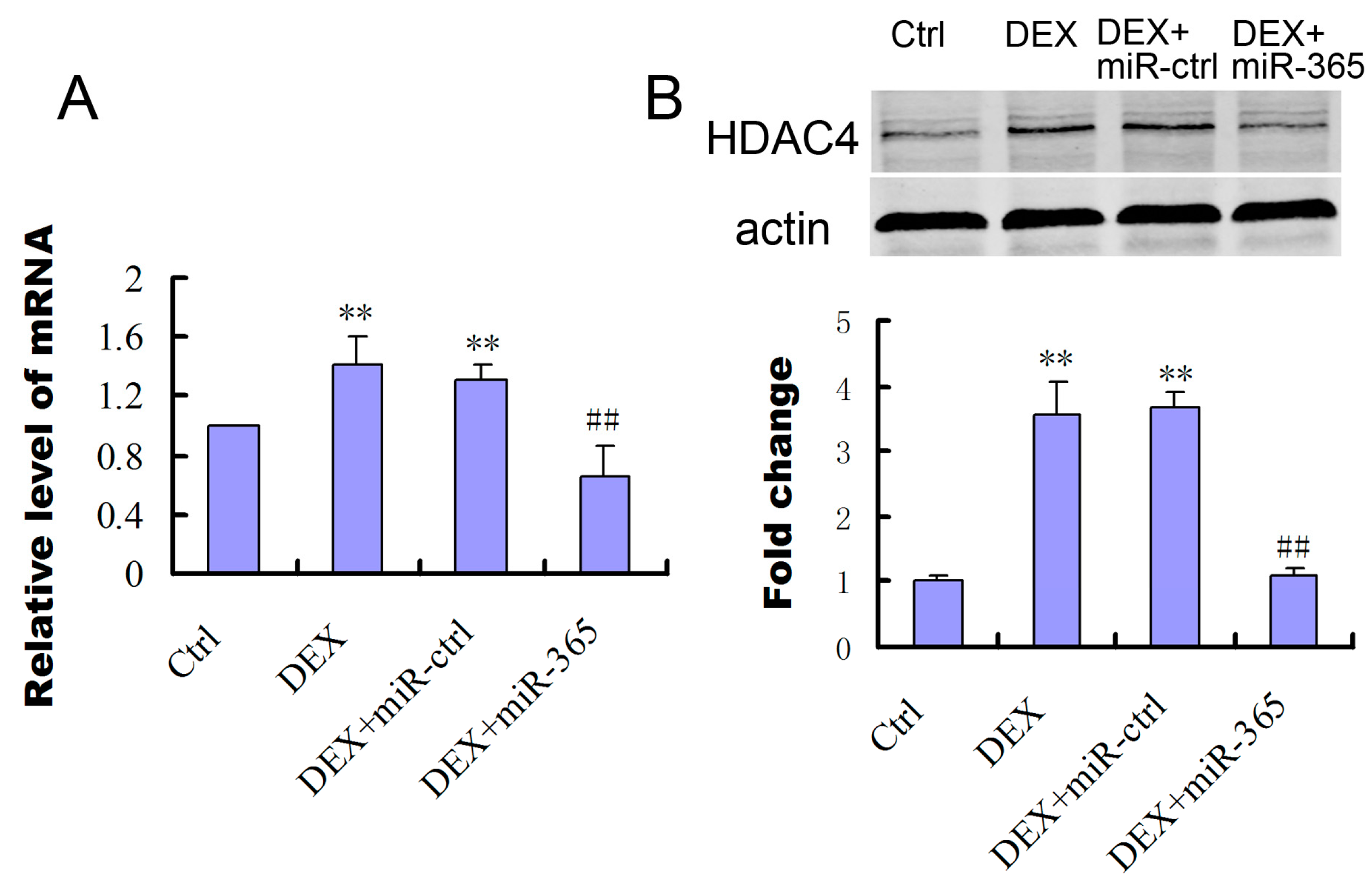
2.4. MiR-365 Over-Expression Inhibited the Upregulation of HDAC4 Induced by DEX
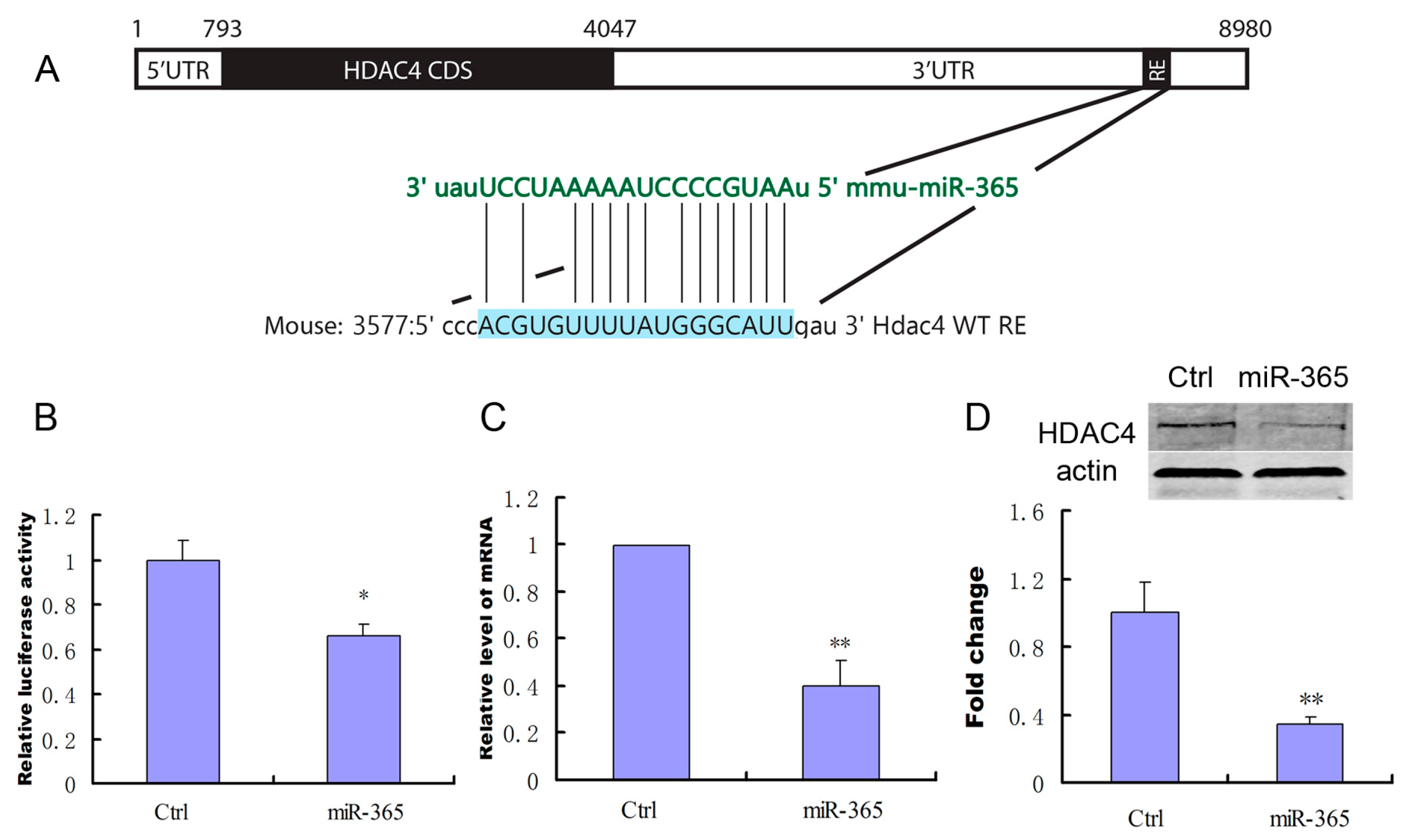
2.5. MiR-365 Directly Targets HDAC4 mRNA in MC3T3-E1 Cells
2.6. MiR-365 Increased Runx2 Expression and HDAC4 Over-Expression Inhibited this Effect
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. MiRNA Transfection
4.3. Cell Viability Assay
4.4. ALP Staining
4.5. RNA Extraction and Real-Time Quantitative PCR
4.6. Luciferase Assays
4.7. Western Blot
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rachner, T.D.; Khosla, S.; Hofbauer, L.C. Osteoporosis: Now and the future. Lancet 2011, 377, 1276–1287. [Google Scholar] [CrossRef]
- Seibel, M.J.; Cooper, M.S.; Zhou, H. Glucocorticoid-induced osteoporosis: Mechanisms, management, and future perspectives. Lancet Diabetes Endocrinol. 2013, 1, 59–70. [Google Scholar] [CrossRef]
- Van der Goes, M.C.; Jacobs, J.W.; Bijlsma, J.W. The value of glucocorticoid co-therapy in different rheumatic diseases—Positive and adverse effects. Arthritis Res. Ther. 2014, 16, S2. [Google Scholar] [CrossRef] [PubMed]
- Rizzoli, R.; Biver, E. Glucocorticoid-induced osteoporosis: Who to treat with what agent? Nat. Rev. Rheumatol. 2015, 11, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S. Glucocorticoid-induced bone disease. N. Engl. J. Med. 2011, 365, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wu, C.; Dong, Y.; Ma, Y.; Jin, Y.; Ji, Y. MicroRNA-24 Regulates Osteogenic Differentiation via Targeting T-Cell Factor-1. Int. J. Mol. Sci. 2015, 16, 11699–11712. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Chen, Y.; Guo, L.; Jiang, T.; Lin, Z. miR-23a/b regulates the balance between osteoblast and adipocyte differentiation in bone marrow mesenchymal stem cells. Bone Res. 2016, 4, 16022. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Dong, Y.; Wu, C.; Ma, Y.; Jin, Y.; Ji, Y. MiR-21 overexpression improves osteoporosis by targeting RECK. Mol. Cell. Biochem. 2015, 405, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Krzeszinski, J.Y.; Wei, W.; Huynh, H.; Jin, Z.; Wang, X.; Chang, T.C.; Xie, X.J.; He, L.; Mangala, L.S.; Lopez-Berestein, G.; et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature 2014, 512, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guan, Y.; Tian, S.; Wang, Y.; Sun, K.; Chen, Q. Mechanical and IL-1β Responsive miR-365 Contributes to Osteoarthritis Development by Targeting Histone Deacetylase 4. Int. J. Mol. Sci. 2016, 17, 436. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.J.; Yang, X.; Wei, L.; Chen, Q. MiR-365: A mechanosensitive microRNA stimulates chondrocyte differentiation through targeting histone deacetylase 4. FASEB J. 2011, 25, 4457–4466. [Google Scholar] [CrossRef] [PubMed]
- Komori, T.; Yagi, H.; Nomura, S.; Yamaguchi, A.; Sasaki, K.; Deguchi, K.; Shimizu, Y.; Bronson, R.T.; Gao, Y.; Inada, M.; et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 1997, 89, 755–764. [Google Scholar] [CrossRef]
- Teitelbaum, S.L. Bone: The conundrum of glucocorticoid-induced osteoporosis. Nat. Rev. Endocrinol. 2012, 8, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S. Glucocorticoid-induced osteoporosis. Rev. Endocr. Metab. Disord. 2001, 2, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Ushijima, K.; Hayashi, Y.; Maekawa, T.; Ando, H.; Tsuruoka, S.; Fujimura, A. Dosing-time dependent effect of dexamethasone on bone density in rats. Life Sci. 2010, 86, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cui, Y.; Chen, Y.; Gao, X.; Su, Y.; Cui, L. Effects of dexamethasone, celecoxib, and methotrexate on the histology and metabolism of bone tissue in healthy Sprague Dawley rats. Clin. Interv. Aging 2015, 10, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Haberland, M.; Montgomery, R.L.; Olson, E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat. Rev. Genet. 2009, 10, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug. Discov. 2012, 11, 384–400. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.A.; Stein, J.L.; Westendorf, J.J.; van Wijnen, A.J. Chromatin modifiers and histone modifications in bone formation, regeneration, and therapeutic intervention for bone-related disease. Bone 2015, 81, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, V.; Tirino, V.; Papaccio, G.; Paino, F. Bone defects: Molecular and cellular therapeutic targets. Int. J. Biochem. Cell Biol. 2014, 51, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Vega, R.B.; Matsuda, K.; Oh, J.; Barbosa, A.C.; Yang, X.; Meadows, E.; McAnally, J.; Pomajzl, C.; Shelton, J.M.; Richardson, J.A.; et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell 2004, 119, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, E.; Selvamurugan, N.; Westendorf, J.J.; Olson, E.N.; Partridge, N.C. HDAC4 represses matrix metalloproteinase-13 transcription in osteoblastic cells, and parathyroid hormone controls this repression. J. Biol. Chem. 2010, 285, 9616–9626. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, T.M.; Westendorf, J.J. Histone deacetylase inhibitors promote osteoblast maturation. J. Bone Miner. Res. 2005, 20, 2254–2263. [Google Scholar] [CrossRef] [PubMed]
- Dudakovic, A.; Evans, J.M.; Li, Y.; Middha, S.; McGee-Lawrence, M.E.; van Wijnen, A.J.; Westendorf, J.J. Histone deacetylase inhibition promotes osteoblast maturation by altering the histone H4 epigenome and reduces Akt phosphorylation. J. Biol. Chem. 2013, 288, 28783–28791. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.Y.; Chuang, P.C.; Chen, M.W.; Ke, H.C.; Wu, S.L.; Chang, Y.H.; Chen, Y.S.; Wang, F.S. MicroRNA-29a ameliorates glucocorticoid-induced suppression of osteoblast differentiation by regulating β-catenin acetylation. Bone 2013, 57, 468–475. [Google Scholar] [CrossRef] [PubMed]
- Ducy, P.; Zhang, R.; Geoffroy, V.; Ridall, A.L.; Karsenty, G. Osf2/Cbfa1: A transcriptional activator of osteoblast differentiation. Cell 1997, 89, 747–754. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, M.; Xiao, G.; Franceschi, R.T. Gene transfer of the Runx2 transcription factor enhances osteogenic activity of bone marrow stromal cells in vitro and in vivo. Mol. Ther. 2005, 12, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-β through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Wei, L.; Zhang, Z.; Guo, L.; Zhang, C.; Li, Y.; Sun, C.; Sun, X.; Wang, S.; Li, P.; Wei, X. Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: A novel mechanism of human osteoarthritis cartilage degeneration. Arthritis Res. Ther. 2014, 16, 491. [Google Scholar] [PubMed]
- Smith, S.S.; Kessler, C.B.; Shenoy, V.; Rosen, C.J.; Delany, A.M. IGF-I 3′ untranslated region: Strain-specific polymorphisms and motifs regulating IGF-I in osteoblasts. Endocrinology 2013, 154, 253–262. [Google Scholar] [CrossRef] [PubMed]



© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, D.; Gao, Y.; Hu, N.; Wu, L.; Chen, Q. miR-365 Ameliorates Dexamethasone-Induced Suppression of Osteogenesis in MC3T3-E1 Cells by Targeting HDAC4. Int. J. Mol. Sci. 2017, 18, 977. https://doi.org/10.3390/ijms18050977
Xu D, Gao Y, Hu N, Wu L, Chen Q. miR-365 Ameliorates Dexamethasone-Induced Suppression of Osteogenesis in MC3T3-E1 Cells by Targeting HDAC4. International Journal of Molecular Sciences. 2017; 18(5):977. https://doi.org/10.3390/ijms18050977
Chicago/Turabian StyleXu, Daohua, Yun Gao, Nan Hu, Longhuo Wu, and Qian Chen. 2017. "miR-365 Ameliorates Dexamethasone-Induced Suppression of Osteogenesis in MC3T3-E1 Cells by Targeting HDAC4" International Journal of Molecular Sciences 18, no. 5: 977. https://doi.org/10.3390/ijms18050977