Comparative Study of Lectin Domains in Model Species: New Insights into Evolutionary Dynamics
Abstract
:1. Introduction
2. Results and Discussion
2.1. Most Plant Lectin Domains Are Widely Distributed in Arabidopsis, Soybean, Cucumber and Rice
2.2. Domain Organization of Lectin Genes in Arabidopsis, Soybean, Cucumber and Rice
2.3. Phylogenetic Relationships and Biological Significance
2.3.1. Ricin B and GH Domains
2.3.2. Nictaba and F-Box Domains
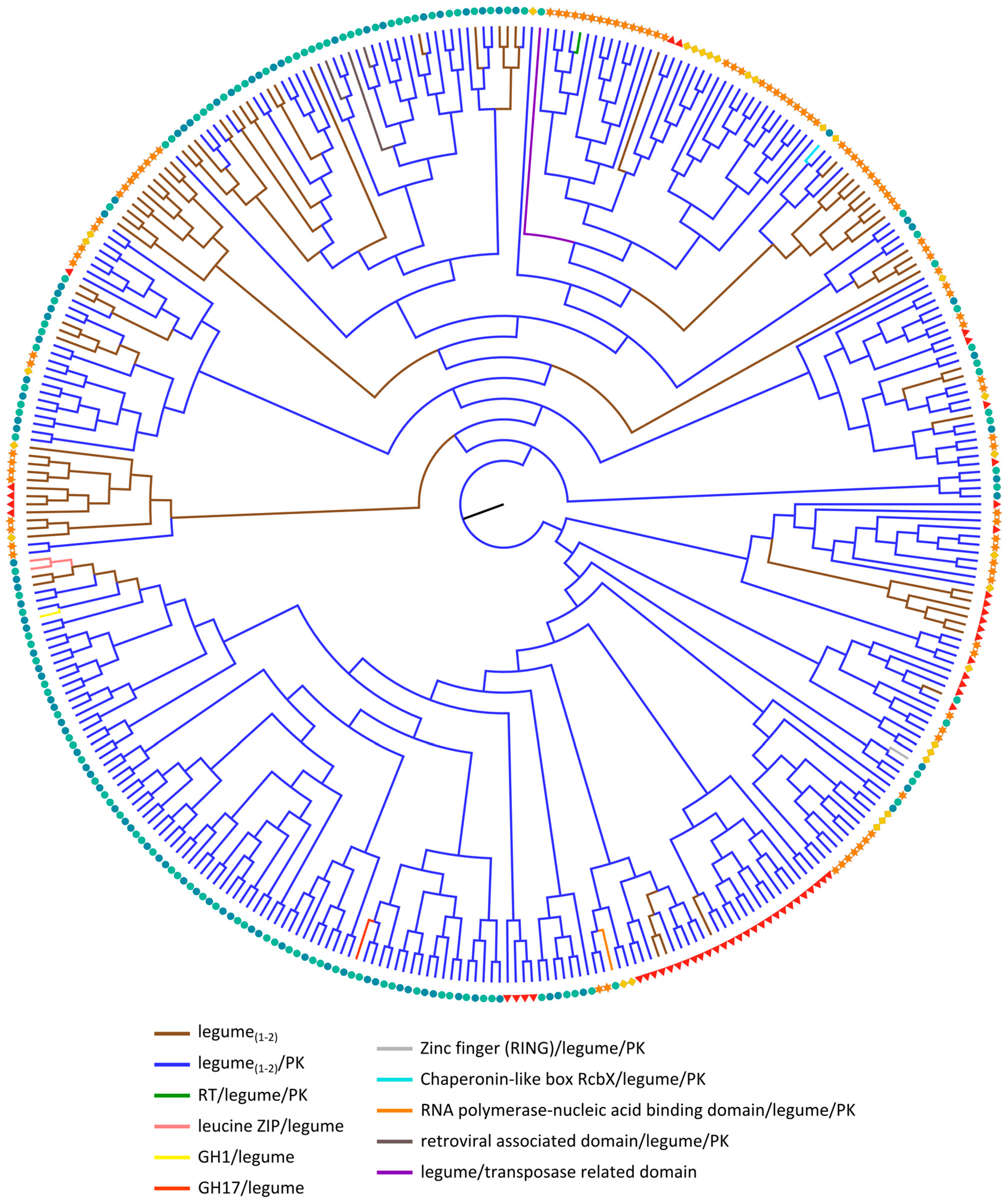
2.3.3. Legume Lectin Homologs
2.3.4. GNA Homologs
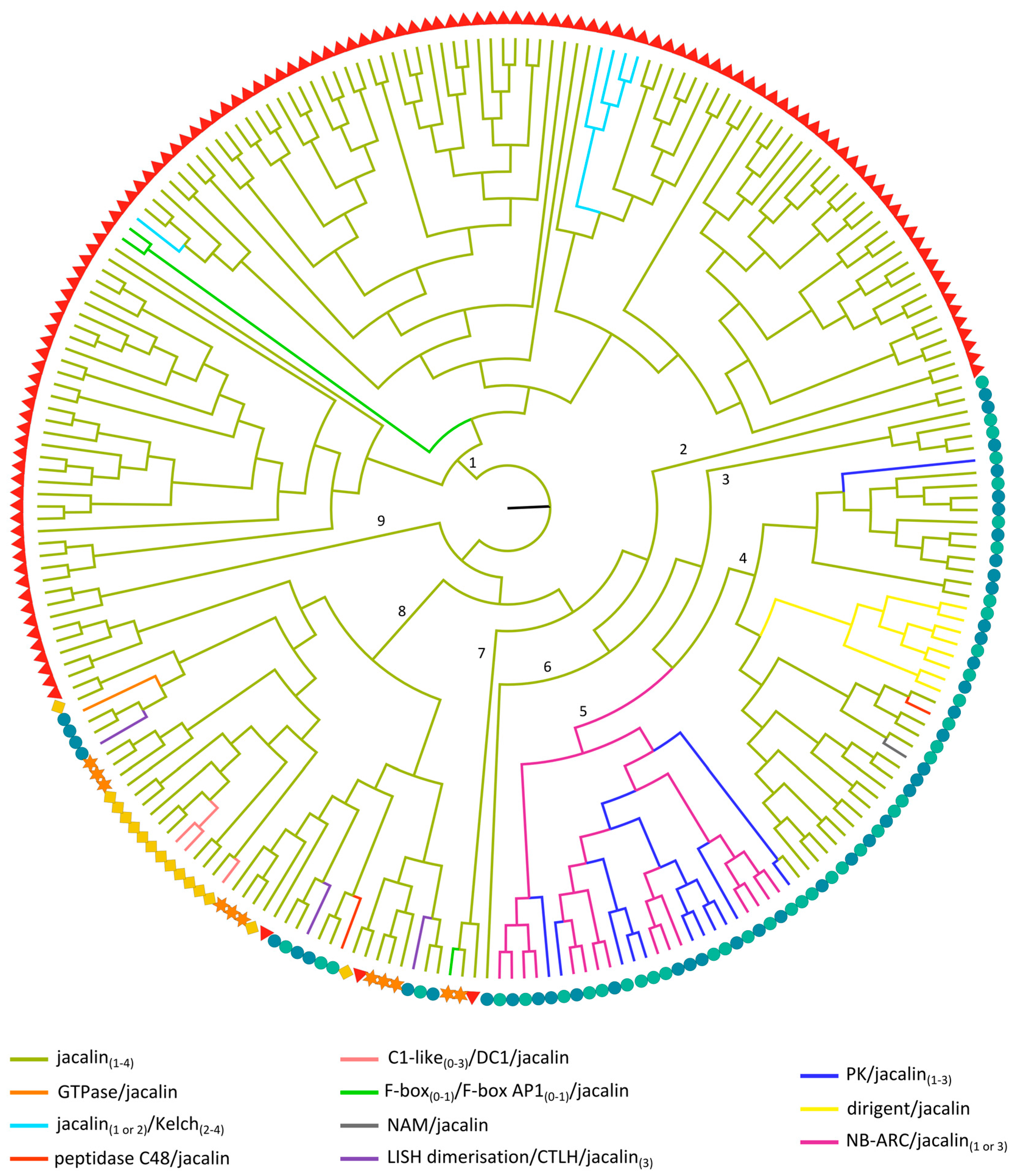
2.3.5. Jacalin-Related Lectin Domains
2.3.6. LysM Domains
2.3.7. Hevein Domains
3. Materials and Methods
3.1. Identification and Classification of Lectin Genes
3.2. Phylogenetics
3.3. Segmental and Tandem Duplications
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Martinez, M. Plant protein-coding gene families: Emerging bioinformatics approaches. Trends Plant Sci. 2011, 16, 558–567. [Google Scholar] [CrossRef] [PubMed]
- Khalturin, K.; Hemmrich, G.; Fraune, S.; Augustin, R.; Bosch, T.C.G. More than just orphans: Are taxonomically-restricted genes important in evolution? Trends Genet. 2009, 25, 404–413. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.N.; Ponting, C.P.; Russell, R.B. On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 2001, 134, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Copley, R.R.; Doerks, T.; Letunic, I.; Bork, P. Protein domain analysis in the era of complete genomes. FEBS Lett. 2002, 513, 129–134. [Google Scholar] [CrossRef]
- Kummerfeld, S.K.; Teichmann, S.A. Relative rates of gene fusion and fission in multi-domain proteins. Trends Genet. 2005, 21, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Weiner, J.; Beaussart, F.; Bornberg-Bauer, E. Domain deletions and substitutions in the modular protein evolution. FEBS J. 2006, 273, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.D.; Björklund, Å.K.; Ekman, D.; Bornberg-Bauer, E.; Elofsson, A. Arrangements in the modular evolution of proteins. Trends Biochem. Sci. 2008, 33, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.D.; Bornberg-Bauer, E. The dynamics and evolutionary potential of domain loss and emergence. Mol. Biol. Evol. 2012, 29, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. USA 2009, 106, 11079–11084. [Google Scholar] [CrossRef] [PubMed]
- Kersting, A.R.; Bornberg-Bauer, E.; Moore, A.D.; Grath, S. Dynamics and adaptive benefits of protein domain emergence and arrangements during plant genome evolution. Genome Biol. Evol. 2012, 4, 316–329. [Google Scholar] [CrossRef] [PubMed]
- Björklund, Å.K.; Ekman, D.; Light, S.; Frey-Skött, J.; Elofsson, A. Domain rearrangements in protein evolution. J. Mol. Biol. 2005, 353, 911–923. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.; Teichmann, S.A.; Pereira-Leal, J. The relationship between domain duplication and recombination. J. Mol. Biol. 2005, 346, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Bashton, M.; Chothia, C. The generation of new protein functions by the combination of domains. Structure 2007, 15, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Van Damme, E.J.M.; Peumans, W.J.; Barre, A.; Rougé, P. Plant lectins: A composite of several distinct families of structurally and evolutionary related proteins with diverse biological roles. CRC Crit. Rev. Plant Sci. 1998, 17, 575–692. [Google Scholar] [CrossRef]
- Van Damme, E.J.M.; Lannoo, N.; Peumans, W.J. Plant lectins. Adv. Bot. Res. 2008, 48, 107–209. [Google Scholar]
- Fouquaert, E.; Smith, D.F.; Peumans, W.J.; Proost, P.; Balzarini, J.; Savvides, S.N.; Van Damme, E.J.M. Related lectins from snowdrop and maize differ in their carbohydrate-binding specificity. Biochem. Biophys. Res. Commun. 2009, 380, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Fouquaert, E.; Van Damme, E.J.M. Promiscuity of the Euonymus carbohydrate-binding domain. Biomolecules 2012, 2, 415–434. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, K.; Lannoo, N.; Proost, P.; Van Damme, E.J.M. Arabidopsis F-box protein containing a Nictaba-related lectin domain interacts with N-acetyllactosamine structures. FEBS Open Bio 2012, 2, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Van Damme, E.J.M. Lectins as plant defense proteins. Plant Physiol. 1995, 109, 347–352. [Google Scholar] [CrossRef] [PubMed]
- Van Hove, J.; De Jaeger, G.; De Winne, N.; Guisez, Y.; Van Damme, E.J.M. The Arabidopsis lectin EULS3 is involved in stomatal closure. Plant Sci. 2015, 238, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Song, M.; Wei, Z.; Tong, J.; Zhang, L.; Xiao, L.; Ma, Z.; Wang, Y. A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 2011, 62, 5471–5483. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Yu, Q.Y.; Tang, L.L.; Ji, W.; Bai, X.; Cai, H.; Liu, X.F.; Ding, X.D.; Zhu, Y.M. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J. Plant Physiol. 2013, 170, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Weide, R.; Govers, F.; Bouwmeester, K. L-type lectin receptor kinases in Nicotiana benthamiana and tomato and their role in Phytophthora resistance. J. Exp. Bot. 2015, 66, 6731–6743. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Lee, D.H.; Choi, D.S.; Hwang, B.K. The pepper GNA-related lectin and PAN domain protein gene, CaGLP1, is required for plant cell death and defense signaling during bacterial infection. Plant Sci. 2015, 241, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Hofberger, J.A.; Nsibo, D.L.; Govers, F.; Bouwmeester, K.; Schranz, M.E. A complex interplay of tandem- and whole-genome duplication drives expansion of the L-type lectin receptor kinase gene family in the Brassicaceae. Genome Biol. Evol. 2015, 7, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Saeed, B.; Baranwal, V.K.; Khurana, P. Identification and expression profiling of the lectin gene superfamily in mulberry. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Sabotič, J.; Ohm, R.A.; Künzler, M. Entomotoxic and nematotoxic lectins and protease inhibitors from fungal fruiting bodies. Appl. Microbiol. Biotechnol. 2016, 100, 91–111. [Google Scholar] [CrossRef] [PubMed]
- Van Holle, S.; Rougé, P.; Van Damme, E.J.M. Evolution and structural diversification of Nictaba-like lectin genes in food crops with a focus on soybean (Glycine max). Ann. Bot. 2017, 119, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Pandey, A.K.; Singh, K.; Upadhyay, S.K. Molecular characterization and global expression analysis of lectin receptor kinases in bread wheat (Triticum aestivum). PLoS ONE 2016, 11, e0153925. [Google Scholar]
- Yang, Y.; Labbé, J.; Muchero, W.; Yang, X.; Jawdy, S.S.; Kennedy, M.; Johnson, J.; Sreedasyam, A.; Schmutz, J.; Tuskan, G.A.; Chen, J.G. Genome-wide analysis of lectin receptor-like kinases in Populus. BMC Genom. 2016, 17, 699. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Li, M.; Liu, P. Evolution of S-domain receptor-like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol. Biol. 2013, 13, 69. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Xu, W.; Xiang, Y.; Jia, H.; Zhang, L.; Ma, Z. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014, 84, 95–110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.C.; Wu, X.; Findley, S.; Wan, J.; Libault, M.; Nguyen, H.T.; Cannon, S.B.; Stacey, G. Molecular evolution of LysM type receptor-like kinases in plants. Plant Physiol. 2007, 144, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.H. Monocot chimeric jacalins: A novel subfamily of plant lectins. Crit. Rev. Biotechnol. 2014, 34, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Van Holle, S.; Van Damme, E.J.M. Distribution and evolution of the lectin family in soybean (Glycine max). Molecules 2015, 20, 2868–2891. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; Van Damme, E.J.M. Genome-wide identification and domain organization of lectin domains in cucumber. Plant Physiol. Biochem. 2016, 108, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Eggermont, L.; Verstraeten, B.; Van Damme, E.J.M. Genome wide screening for lectin motifs in Arabidopsis thaliana. Plant Genome 2017. [Google Scholar] [CrossRef]
- Percudani, R.; Montanini, B.; Ottonello, S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins 2005, 60, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Fouquaert, E.; Jauneau, A.; Rougé, P.; Lannoo, N.; Hamada, H.; Alvarez, R.; Devreese, B.; Van Damme, E.J.M. The liverwort Marchantia polymorpha expresses orthologs of the fungal Agaricus bisporus agglutinin family. Plant Physiol. 2007, 144, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Bovi, M.; Carrizo, M.E.; Capaldi, S.; Perduca, M.; Chiarelli, L.R.; Galliano, M.; Monaco, H.L. Structure of a lectin with antitumoral properties in king bolete (Boletus edulis) mushrooms. Glycobiology 2011, 21, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Dang, L. Chimeric Lectins with Toxin Domains from Cucumber. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2017. [Google Scholar]
- Faruque, K.; Begam, R.; Deyholos, M.K. The Amaranthin-like lectin (LuALL) genes of flax: A unique gene family with members inducible by defence hormones. Plant Mol. Biol. Rep. 2015, 33, 731–741. [Google Scholar] [CrossRef]
- Sémon, M.; Wolfe, K.H. Consequences of genome duplication. Curr. Opin. Genet. Dev. 2007, 17, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F. Genome evolution in polyploids. Plant Mol. Biol. 2000, 42, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Schranz, E.M.; Mohammadin, S.; Edger, P.P. Ancient whole genome duplications, novelty and diversification: The WGD radiation lag-time model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Van de Peer, Y.; Maere, S.; Meyer, A. The evolutionary significance of ancient genome duplications. Nat. Rev. Genet. 2009, 10, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Hokamp, K.; Wolfe, K.H. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003, 13, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.G.; Xiong, Y.Q.; Liu, T.Y.; Sun, S.H.; Chen, L.; Chen, M.S. Evidence for an ancient whole-genome duplication event in rice and other cereals. Yi Chuan Xue Bao 2005, 32, 519–527. [Google Scholar] [PubMed]
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C.; et al. The genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3, e38. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Chapman, B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 2004, 101, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, R.C.; Schlueter, J.; Doyle, J.J. Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 2006, 9, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Combes, M.C.; Lashermes, P. Comparative sequence analyses indicate that Coffea (Asterids) and Vitis (Rosids) derive from the same paleo-hexaploid ancestral genome. Mol. Genet. Genom. 2010, 283, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Proost, S.; Van Bel, M.; Vaneechoutte, D.; Van de Peer, Y.; Inzé, D.; Mueller-Roeber, B.; Vandepoele, K. PLAZA 3.0: An access point for plant comparative genomics. Nucleic Acids Res. 2015, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.C.H.; Blumwald, E. Domains as functional building blocks of plant proteins. Trends Plant Sci. 2002, 7, 544–549. [Google Scholar] [CrossRef]
- Fouquaert, E.; Peumans, W.J.; Vandekerckhove, T.T.M.; Ongenaert, M.; Van Damme, E.J.M. Proteins with an Euonymus lectin-like domain are ubiquitous in Embryophyta. BMC Plant Biol. 2009, 9, 136. [Google Scholar] [CrossRef] [PubMed]
- De Schutter, K.; Tsaneva, M.; Kulkarni, S.R.; Rougé, P.; Vandepoele, K.; Van Damme, E.J.M. Evolutionary relationships and expression analysis of EUL domain proteins in rice (Oryza sativa). Rice 2017. [Google Scholar] [CrossRef]
- Zhang, X.C.; Wang, Z.; Zhang, X.; Le, M.H.; Sun, J.; Xu, D.; Cheng, J.; Stacey, G. Evolutionary dynamics of protein domain architecture in plants. BMC Evol. Biol. 2012, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Van Damme, E.J.M. Evolution of plant ribosome-inactivating proteins. In Plant Cell Monographs; Lord, J.M., Hartley, R.M., Eds.; Springer: New York, NY, USA, 2010; Volume 18, pp. 1–26. [Google Scholar]
- Aspeborg, H.; Coutinho, P.M.; Wang, Y.; Brumer, H.; Henrissat, B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol. Biol. 2012, 12, 186. [Google Scholar] [CrossRef] [PubMed]
- Olsnes, S. The history of ricin, abrin and related toxins. Toxicon 2004, 44, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, L.; Valbonesi, P.; Gorini, P.; Pession, A.; Stirpe, F. Polynucleotide: Adenosine glycosidase activity of saporin-L1: effect on DNA, RNA and poly (A). Biochem. J. 1996, 319, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Bolognesi, A.; Bortolotti, M.; Maiello, S.; Battelli, M.; Polito, L. Ribosome-inactivating proteins from plants: A historical overview. Molecules 2016, 21, 1627. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Etzler, M.E. R-type Lectins. In Essentials of Glycobiology, 2nd ed.; Varki, A., Cummings, R., Esko, J., Freeze, H., Stanley, P., Bertozzi, C., Hart, G., Etzler, M., Eds.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2009; pp. 403–414. [Google Scholar]
- Vandenbussche, F.; Peumans, W.J.; Desmyter, S.; Proost, P.; Ciani, M.; Van Damme, E.J.M. The type-1 and type-2 ribosome-inactivating proteins from Iris confer transgenic tobacco plants local but not systemic protection against viruses. Planta 2004, 220, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Shahidi-Noghabi, S.; Van Damme, E.J.M.; Smagghe, G. Expression of Sambucus nigra agglutinin (SNA-I) from elderberry bark in transgenic tobacco plants results in enhanced resistance to different insect species. Transgenic Res. 2009, 18, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Wei, G.Q.; Liu, R.S.; Wang, Q.; Liu, W.Y. Toxicity of two type II ribosome-inactivating proteins (cinnamomin and ricin) to domestic silkworm larvae. Arch. Insect Biochem. Physiol. 2004, 57, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Schrot, J.; Weng, A.; Melzig, M.F. Ribosome-inactivating and related proteins. Toxins 2015, 7, 1556–1615. [Google Scholar] [CrossRef] [PubMed]
- Babu, M.; Griffiths, J.S.; Huang, T.S.; Wang, A. Altered gene expression changes in Arabidopsis leaf tissues and protoplasts in response to Plum pox virus infection. BMC Genom. 2008, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Sakuta, C.; Masuda, S.; Mizoguchi, T.; Kamada, H.; Satoh, S. Possible involvement of leaf gibberellins in the clock-controlled expression of XSP30, a gene encoding a xylem sap lectin, in cucumber roots. Plant Physiol. 2003, 133, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Kittur, F.S.; Mangala, S.L.; Rus’d, A.A.; Kitaoka, M.; Tsujibo, H.; Hayashi, K. Fusion of family 2b carbohydrate-binding module increases the catalytic activity of a xylanase from Thermotoga maritima to soluble xylan. FEBS Lett. 2003, 549, 147–151. [Google Scholar] [CrossRef]
- Mangala, S.L.; Kittur, F.S.; Nishimoto, M.; Sakka, K.; Ohmiya, K.; Kitaoka, M.; Hayashi, K. Fusion of family VI cellulose binding domains to Bacillus halodurans xylanase increases its catalytic activity and substrate-binding capacity to insoluble xylan. J. Mol. Catal. B. Enzym. 2003, 21, 221–230. [Google Scholar] [CrossRef]
- Fujimoto, Z. Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a β-trefoil fold. Biosci. Biotechnol. Biochem. 2013, 77, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Ma, H.; Nei, M.; Kong, H. Evolution of F-box genes in plants: Different modes of sequence divergence and their relationships. Proc. Natl. Acad. Sci. USA 2009, 106, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Peumans, W.J.; Hause, B.; Bras, J.; Kumar, M.; Proost, P.; Barre, A.; Rougé, P.; Van Damme, E.J. M. Jasmonate methyl ester induces the synthesis of a cytoplasmic/nuclear chitooligosaccharide-binding lectin in tobacco leaves. FASEB J. 2002, 16, 905–907. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Vandenborre, G.; Miersch, O.; Smagghe, G.; Wasternack, C.; Peumans, W.J.; Van Damme, E.J. M. The jasmonate-induced expression of the Nicotiana tabacum leaf lectin. Plant Cell Physiol. 2007, 48, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Vandenborre, G.; Van Damme, E.J. M.; Smagghe, G. Nicotiana tabacum agglutinin expression in response to different biotic challengers. Arthropod. Plant Interact. 2009, 3, 193–202. [Google Scholar] [CrossRef]
- Van Holle, S.; Smagghe, G.; Van Damme, E.J.M. Overexpression of Nictaba-like lectin genes from Glycine max confers tolerance toward Pseudomonas syringae infection, aphid infestation and salt stress in transgenic Arabidopsis plants. Front Plant Sci. 2016, 7, 1590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shi, H.; Chen, L.; Wang, X.; Lü, B.; Zhang, S.; Liang, Y.; Liu, R.; Qian, J.; Sun, W.; You, Z.; Dong, H. Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 2011, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Beneteau, J.; Renard, D.; Marché, L.; Douville, E.; Lavenant, L.; Rahbé, Y.; Dupont, D.; Vilaine, F.; Dinant, S. Binding properties of the N-acetylglucosamine and high-mannose N-glycan PP2-A1 phloem lectin in Arabidopsis. Plant Physiol. 2010, 153, 1345–1361. [Google Scholar] [CrossRef] [PubMed]
- Delporte, A.; De Vos, W.H.; Van Damme, E.J. M. In vivo interaction between the tobacco lectin and the core histone proteins. J. Plant Physiol. 2014, 171, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Reuber, T.L.; Ausubel, F.M. Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease resistance genes. Plant Cell 1996, 8, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Ve, T.; Williams, S.J.; Kobe, B. Structure and function of Toll/interleukin-1 receptor/resistance protein (TIR) domains. Apoptosis 2015, 20, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Bai, C.; Sen, P.; Hofmann, K.; Ma, L.; Goebl, M.; Harper, J.; Elledge, S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996, 86, 263–274. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; Kuroda, H.; Kuromori, T.; Hirayama, T.; Seki, M.; Shinozaki, K.; Shimada, H.; Matsui, M. Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol. 2004, 45, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Kipreos, E.T.; Pagano, M. The F-box protein family. Genome Biol. 2000, 1, reviews3002. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, T.; Yoshida, Y.; Kumanomidou, T.; Hasegawa, Y.; Suzuki, A.; Yamane, T.; Tanaka, K. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2007, 104, 5777–5781. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, H.; Yanagawa, Y.; Takahashi, N.; Horii, Y.; Matsui, M. A comprehensive analysis of interaction and localization of Arabidopsis SKP1-LIKE (ASK) and F-Box (FBX) proteins. PLoS ONE 2012, 7, e50009. [Google Scholar] [CrossRef] [PubMed]
- Narsai, R.; Wang, C.; Chen, J.; Wu, J.; Shou, H.; Whelan, J. Antagonistic, overlapping and distinct responses to biotic stress in rice (Oryza sativa) and interactions with abiotic stress. BMC Genom. 2013, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Stefanowicz, K.; Lannoo, N.; Zhao, Y.; Eggermont, L.; Van Hove, J.; Al Atalah, B.; Van Damme, E.J.M. Glycan-binding F-box protein from Arabidopsis thaliana protects plants from Pseudomonas syringae infection. BMC Plant Biol. 2016, 16, 213. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Wang, C.; Huang, J.; Yang, G.; Wu, C.; Zheng, C. SCF E3 ligase PP2-B11 plays a positive role in response to salt stress in Arabidopsis. J. Exp. Bot. 2015, 66, 4683–4697. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Govers, F. Arabidopsis L-type lectin receptor kinases: Phylogeny, classification, and expression profiles. J. Exp. Bot. 2009, 60, 4383–4396. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Gochicoa, M.T.; Camut, S.; Timmers, A.C. J.; Niebel, A.; Hervé, C.; Boutet, E.; Bono, J.-J.; Imbery, A.; Cullimore, J.V. Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula. Structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol. 2003, 133, 1893–1910. [Google Scholar] [CrossRef] [PubMed]
- Brewin, N.J.; Kardailsky, I.V. Legume lectins and nodulation by Rhizobium. Trends Plant Sci. 1997, 2, 92–98. [Google Scholar] [CrossRef]
- Frenzel, A.; Tiller, N.; Hause, B.; Krajinski, F. The conserved arbuscular mycorrhiza-specific transcription of the secretory lectin MtLec5 is mediated by a short upstream sequence containing specific protein binding sites. Planta 2006, 224, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cordewener, J.H.G.; America, A.H.P.; Shan, W.X.; Bouwmeester, K.; Govers, F. Arabidopsis lectin receptor kinases LecRK-IX.1 and LecRK-IX.2 are functional analogs in regulating Phytophthora resistance and plant cell death. Mol. Plant Microbe Interact. 2015, 28, 1032–1048. [Google Scholar] [CrossRef] [PubMed]
- Bouwmeester, K.; Han, M.; Blanco-Portales, R.; Song, W.; Weide, R.; Guo, L.Y.; van der Vossen, E.A.G.; Govers, F. The Arabidopsis lectin receptor kinase LecRK-I.9 enhances resistance to Phytophthora infestans in Solanaceous plants. Plant Biotechnol. J. 2014, 12, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, H.; Saitoh, H.; Takahashi, Y.; Berberich, T.; Ito, A.; Kamoun, S.; Terauchi, R. NbLRK1, a lectin-like receptor kinase protein of Nicotiana benthamiana, interacts with Phytophthora infestans INF1 elicitin and mediates INF1-induced cell death. Planta 2008, 228, 977–987. [Google Scholar] [CrossRef] [PubMed]
- Gouget, A.; Senchou, V.; Govers, F.; Sanson, A.; Barre, A.; Rougé, P.; Pont-Lezica, R.; Canut, H. Lectin receptor kinases participate in protein-protein interactions to mediate plasma membrane-cell wall adhesions in Arabidopsis. Plant Physiol. 2006, 140, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Wang, R.; Xu, X.; He, X.; Sun, B.; Zhong, Y.; Liang, Z.; Luo, S.; Lin, Y. Cucumis sativus L-type lectin receptor kinase (CsLecRK) gene family response to Phytophthora melonis, Phytophthora capsici and water immersion in disease resistant and susceptible cucumber cultivars. Gene 2014, 549, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Ju, H.W.; Min, J.H.; Zhang, X.; Kim, S.H.; Yang, K.Y.; Kim, C.S. Overexpression of L-type lectin-like protein kinase 1 confers pathogen resistance and regulates salinity response in Arabidopsis thaliana. Plant Sci. 2013, 203–204, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Gilardoni, P.A.; Hettenhausen, C.; Baldwin, I.T.; Bonaventure, G. Nicotiana attenuata LECTIN RECEPTOR KINASE1 suppresses the insect-mediated inhibition of induced defense responses during Manduca sexta herbivory. Plant Cell 2011, 23, 3512–3532. [Google Scholar] [CrossRef] [PubMed]
- Desclos-Theveniau, M.; Arnaud, D.; Huang, T.Y.; Lin, G.J. C.; Chen, W.Y.; Lin, Y.C.; Zimmerli, L. The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 2012, 8, e1002513. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Kuo, Y.C.; Mishra, S.; Tsai, C.H.; Chien, C.C.; Chen, C.W.; Desclos-Theveniau, M.; Chu, P.W.; Schulze, B.; Chinchilla, D.; et al. The lectin receptor kinase-VI.2 is required for priming and positively regulates Arabidopsis pattern-triggered immunity. Plant Cell 2012, 24, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Vaid, N.; Pandey, P.; Srivastava, V.K.; Tuteja, N. Pea lectin receptor-like kinase functions in salinity adaptation without yield penalty, by alleviating osmotic and ionic stresses and upregulating stress-responsive genes. Plant Mol. Biol. 2015, 88, 193–206. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Tanaka, K.; Nguyen, C.T.; Stacey, G. Extracellular ATP is a central signaling molecule in plant stress responses. Curr. Opin. Plant Biol. 2014, 20, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Shiu, S.; Karlowski, W.; Pan, R. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Kachroo, A.; Schopfer, C.R.; Nasrallah, M.E.; Nasrallah, J.B. Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 2001, 293, 1824–1826. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.T.; Oh, J.; Kim, K.H.; Uhm, J.Y.; Lee, B.M. Isolation and characterization of NgRLK1, a receptor-like kinase of Nicotiana glutinosa that interacts with the elicitin of Phytophthora capsici. Mol. Biol. Rep. 2010, 37, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Lenka, S.K.; Lohia, B.; Kumar, A.; Chinnusamy, V.; Bansal, K.C. Genome-wide targeted prediction of ABA responsive genes in rice based on over-represented cis-motif in co-expressed genes. Plant Mol. Biol. 2009, 69, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Bonaventure, G. The Nicotiana attenuata LECTIN RECEPTOR KINASE 1 is involved in the perception of insect feeding. Plant Signal Behav. 2011, 6, 2060–2063. [Google Scholar] [CrossRef] [PubMed]
- Xu, E.; Vaahtera, L.; Brosché, M. Roles of defense hormones in the regulation of ozone-induced changes in gene expression and cell death. Mol. Plant 2015, 8, 1776–1794. [Google Scholar] [CrossRef] [PubMed]
- Sanabria, N.; Goring, D.; Nürnberger, T.; Dubery, I. Self/nonself perception and recognition mechanisms in plants: A comparison of self-incompatibility and innate immunity. New Phytol. 2008, 178, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Blaum, B.S.; Mazzotta, S.; Nöldeke, E.R.; Halter, T.; Madlung, J.; Kemmerling, B.; Stehle, T. Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 2014, 186, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.J.; Wuriyanghan, H.; Zhang, Y.Q.; Duan, K.X.; Chen, H.W.; Li, Q.T.; Lu, X.; He, S.J.; Ma, B.; Zhang, W.K.; et al. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol. 2013, 163, 1752–1765. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Wu, Y.; Guo, J.; Du, B.; Chen, R.; Zhu, L.; He, G. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013, 76, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wu, H.; Chen, H.; Liu, Y.; He, J.; Kang, H.; Sun, Z.; Pan, G.; Wang, Q.; Hu, J.; et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2014, 33, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shang, J.; Chen, D.; Lei, C.; Zou, Y.; Zhai, W.; Liu, G.; Xu, J.; Ling, Z.; Cao, G.; et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006, 46, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Woloshuk, C.P.; Meulenhoff, J.S.; Sela-Buurlage, M.B.; van den Elzen, P.J.M.; Cornelissen, B.J.C. Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell 1991, 3, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Schaper, E.; Anisimova, M. The evolution and function of protein tandem repeats in plants. New Phytol. 2015, 206, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Jones, J.D.G. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 575–607. [Google Scholar] [CrossRef] [PubMed]
- Peumans, W.J.; Barre, A.; Bras, J.; Rougé, P.; Proost, P.; Van Damme, E.J.M. The liverwort contains a lectin that is structurally and evolutionary related to the monocot mannose-binding lectins. Plant Physiol. 2002, 129, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Lannoo, N.; Van Damme, E.J.M. Lectin domains at the frontiers of plant defense. Front Plant Sci. 2014, 5, 397. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Li, J.; Xu, Y.; Xu, Z.; Chong, K. Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS ONE 2009, 4, e4854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Peumans, W.J.; Barre, A.; Astoul, C.H.; Rovira, P.; Rougé, P.; Proost, P.; Truffa-Bachi, P.; Jalali, A.A.; Van Damme, E.J.M. Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 2000, 210, 970–978. [Google Scholar] [PubMed]
- Al Atalah, B.; Smagghe, G.; Van Damme, E.J.M. Orysata, a jacalin-related lectin from rice, could protect plants against biting-chewing and piercing-sucking insects. Plant Sci. 2014, 221–222, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.M.; Zhang, Q.; Zhao, W.S.; Wang, Y.Y.; Peng, Y.L. Identification of a lectin gene induced in rice in response to Magnaporthe grisea infection. Acta. Bot. Sin. 2003, 23, 799–810. [Google Scholar]
- De Souza Filho, G.A.; Ferreira, B.S.; Dias, J.M.; Queiroz, K.S.; Branco, A.T.; Bressan-Smith, R.E.; Oliveira, J.G.; Garcia, A.B. Accumulation of SALT protein in rice plants as a response to environmental stresses. Plant Sci. 2003, 164, 623–628. [Google Scholar] [CrossRef]
- Patishtan, J.; Hartley, T.N.; Fonseca de Carvalho, R.; Maathuis, F.J. Genome wide association studies to identify rice salt-tolerance markers. Plant Cell Environ 2017, in press. [Google Scholar]
- He, X.; Li, L.; Xu, H.; Xi, J.; Cao, X.; Xu, H.; Rong, S.; Dong, Y.; Wang, C.; Chen, R.; et al. A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. Plant Biol. 2017, 19, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Shinjo, A.; Araki, Y.; Hirano, K.; Arie, T.; Ugaki, M.; Teraoka, T. Transgenic rice plants that over-express the mannose-binding rice lectin have enhanced resistance to rice blast. J. Gen. Plant Pathol. 2011, 77, 85–92. [Google Scholar] [CrossRef]
- Yamaji, Y.; Maejima, K.; Ozeki, J.; Komatsu, K.; Shiraishi, T.; Okano, Y.; Himeno, M.; Sugawara, K.; Neriya, Y.; Minato, N.; et al. Lectin-mediated resistance impairs plant virus infection at the cellular level. Plant Cell 2012, 24, 778–793. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Li, C.; Xu, S.; Xing, L.; Xu, Y.; Chong, K. AtJAC1 regulates nuclear accumulation of GRP7, influencing RNA processing of FLC antisense transcripts and flowering time in Arabidopsis. Plant Physiol. 2015, 169, 2102–2117. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Parra, M.A.; Anderberg, R.J.; Carrington, J.C. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 2001, 127, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Revers, F.; Guiraud, T.; Houvenaghel, M.-C.; Mauduit, T.; Le Gall, O.; Candresse, T. Multiple resistance phenotypes to lettuce mosaic virus among Arabidopsis thaliana accessions. MPMI Mol. Plant Microbe Interact. 2003, 16, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Decroocq, V.; Sicard, O.; Alamillo, J.M.; Lansac, M.; Eyquard, J.P.; García, J.A.; Candresse, T.; Le Gall, O.; Revers, F. Multiple resistance traits control Plum pox virus infection in Arabidopsis thaliana. MPMI Mol. Plant Microbe Interact. 2006, 19, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Van der Biezen, E.A.; Jones, J.D.G. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R228. [Google Scholar] [CrossRef]
- Johnson, J.E.; Goulding, R.E.; Ding, Z.; Partovi, A.; Anthony, K.V.; Beaulieu, N.; Tazmini, G.; Cornell, R.B.; Kay, R.J. Differential membrane binding and diacylglycerol recognition by C1 domains of RasGRPs. Biochem. J. 2007, 406, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Valster, A.H.; Hepler, P.K.; Chernoff, J. Plant GTPases: The Rhos in bloom. Trends Cell Biol. 2000, 10, 141–146. [Google Scholar] [CrossRef]
- Zhang, X.C.; Cannon, S.B.; Stacey, G. Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 2009, 9, 183. [Google Scholar] [CrossRef] [PubMed]
- Buist, G.; Steen, A.; Kok, J.; Kuipers, O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008, 68, 838–847. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Willmann, R.; Desaki, Y.; Grabherr, H.M.; Nürnberger, T. Plant LysM proteins: Modules mediating symbiosis and immunity. Trends Plant Sci. 2012, 17, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakano, T.; Takamizawa, D.; Desaki, Y.; Ishii-Minami, N.; Nishizawa, Y.; Minami, E.; Okada, K.; Yamane, H.; Kaku, H.; et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010, 64, 204–214. [Google Scholar] [CrossRef] [PubMed]
- Petutschnig, E.K.; Jones, A.M.E.; Serazetdinova, L.; Lipka, U.; Lipka, V. The Lysin Motif Receptor-like Kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 2010, 285, 28902–28911. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; Stougaard, J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Knogge, W.; Scheel, D. LysM receptors recognize friend and foe. Proc. Natl. Acad. Sci. USA 2006, 103, 10829–10830. [Google Scholar] [CrossRef] [PubMed]
- Limpens, E.; Franken, C.; Smit, P.; Willemse, J.; Bisseling, T.; Geurts, R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003, 302, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Indrasumunar, A.; Kereszt, A.; Searle, I.; Miyagi, M.; Li, D.; Nguyen, C.D.T.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.). Plant Cell Physiol. 2010, 51, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Indrasumunar, A.; Searle, I.; Lin, M.H.; Kereszt, A.; Men, A.; Carroll, B.J.; Gresshoff, P.M. Nodulation factor receptor kinase 1α controls nodule organ number in soybean (Glycine max L. Merr). Plant J. 2011, 65, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Amor, B.B.; Shaw, S.L.; Oldroyd, G.E.D.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Dénarié, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Neuhaus, J.M.; Fritig, B.; Linthorst, H.J.M.; Meins, F.; Mikkelsen, J.D.; Ryals, J. A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 1996, 14, 102–104. [Google Scholar] [CrossRef]
- Ludvigsen, S.; Poulsen, F.M. Three-dimensional structure in solution of barwin, a protein from barley seed. Biochemistry 1992, 31, 8783–8789. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Lee, H.I.; Davis, K.R. Ozone-induced ethylene production is dependent on salicylic acid, and both salicylic acid and ethylene act in concert to regulate ozone-induced cell death. Plant J. 2002, 32, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Carp, M.J.; Zuchman, R.; Ziv, T.; Horwitz, B. A.; Gepstein, S. Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J. Proteom. 2010, 73, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Eggermont, K.; Penninckx, I.A.M.A.; Mauch-Mani, B.; Vogelsang, R.; Cammue, B.P.A.; Broekaert, W.F. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 1998, 95, 15107–15111. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, A.K.; Xiang, F.; Park, C.M. Molecular and functional profiling of Arabidopsis pathogenesis-related genes: Insights into their roles in salt response of seed germination. Plant Cell Physiol. 2008, 49, 334–344. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.; Uknes, S.; Lawton, K.; Winter, A.M.; Chandler, D.; DiMaio, J.; Novitzky, R.; Ward, E.; Ryals, J. Regulation of a hevein-like gene in Arabidopsis. MPMI Mol. Plant Microbe Interact. 1993, 6, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Eggermont, K.; Tierens, K.F.; Broekaert, W.F. Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 1999, 121, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- de A Gerhardt, L.B.; Sachetto-Martins, G.; Contarini, M.G.; Sandroni, M.; de P Ferreira, R.; de Lima, V.M.; Cordeiro, M.C.; de Oliveira, D.E.; Margis-Pinheiro, M. Arabidopsis thaliana class IV chitinase is early induced during the interaction with Xanthomonas campestris. FEBS Lett. 1997, 419, 69–75. [Google Scholar] [CrossRef]
- de A Gerhardt, L.B.; Magioli, C.; Perez, A.B.U.C.M.; Margis, R.; Sachetto-Martins, G.; Margis-Pinheiro, M. AtchitIV gene expression is stimulated under abiotic stresses and is spatially and temporally regulated during embryo development. Genet. Mol. Biol. 2004, 27, 118–123. [Google Scholar] [CrossRef]
- Gijzen, M.; Kuflu, K.; Qutob, D.; Chernys, J.T. A class I chitinase from soybean seed coat. J. Exp. Bot. 2001, 52, 2283–2289. [Google Scholar] [CrossRef] [PubMed]
- Bagnaresi, P.; Biselli, C.; Orrù, L.; Urso, S.; Crispino, L.; Abbruscato, P.; Piffanelli, P.; Lupotto, E.; Cattivelli, L.; Valè, G. Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE 2012, 7, e51609. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, Y.; Nishio, Z.; Nakazono, K.; Soma, M.; Nakajima, E.; Ogaki, M.; Hibi, T. Enhanced resistance to blast (Magnaporthe grisea) in transgenic japonica rice by constitutive expression of rice chitinase. Theor. Appl. Genet. 1999, 99, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Kovács, G.; Sági, L.; Jacon, G.; Arinaitwe, G.; Busogoro, J.P.; Thiry, E.; Strosse, H.; Swennen, R.; Remy, S. Expression of a rice chitinase gene in transgenic banana (’Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic Res. 2013, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Wang, J.; Du, Z.; Zhang, C.; Li, L.; Xu, Z. Enhanced resistance to stripe rust disease in transgenic wheat expressing the rice chitinase gene RC24. Transgenic Res. 2013, 22, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, C.; Proost, S.; Hernandez-Coronado, M.; Ortiz-Ramirez, C.; Lang, D.; Rensing, S.A.; Becker, J.D.; Vandepoele, K.; Mutwil, M. Phylogenomic analysis of gene co-expression networks reveals evolution of functional modules. Plant J. 2017, 90, 447–465. [Google Scholar] [CrossRef] [PubMed]
- Ruprecht, C.; Vaid, N.; Proost, S.; Persson, S.; Mutwil, M. Beyond genomics: Studying evolution with gene coexpression networks. Trends Plant Sci. 2017, 22, 298–307. [Google Scholar] [CrossRef] [PubMed]





| Lectin Family | Predicted Genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis Thaliana | Glycine Max | Cucumis Sativus | Oryza Sativa (Japonica) | Oryza Sativa (Indica) | ||||||
| # | % | # | % | # | % | # | % | # | % | |
| Amaranthin | 0 | 0.0 | 0 | 0.0 | 16 | 11.0 | 0 | 0.0 | 0 | 0.0 |
| CRA | 9 | 4.2 | 6 | 1.6 | 4 | 2.7 | 2 | 0.6 | 4 | 1.5 |
| EUL | 1 | 0.5 | 3 | 0.8 | 1 | 0.7 | 5 | 1.5 | 5 | 1.8 |
| GNA | 49 | 22.7 | 166 | 45.1 | 45 | 30.8 | 134 | 41.2 | 94 | 34.3 |
| Hevein | 10 | 4.6 | 6 | 1.6 | 4 | 2.7 | 10 | 3.1 | 10 | 3.6 |
| Jacalin | 50 | 23.1 | 5 | 1.4 | 8 | 5.5 | 30 | 9.2 | 33 | 12.0 |
| Legume lectin | 54 | 25.0 | 94 | 25.5 | 29 | 19.9 | 104 | 32.0 | 84 | 30.7 |
| LysM | 12 | 5.6 | 47 | 12.8 | 10 | 6.8 | 20 | 6.2 | 22 | 8.0 |
| Nictaba | 29 | 13.4 | 31 | 8.4 | 20 | 13.7 | 20 | 6.2 | 22 | 8.0 |
| Ricin B | 2 | 0.9 | 10 | 2.7 | 9 | 6.2 | 4 | 1.2 | 4 | 1.4 |
| Total number of lectin sequences | 216 | 368 | 146 | 329 | 278 | |||||
| Domain Architecture | At | Gm | Cs | Os ja | Os in | Schematic Representation |
|---|---|---|---|---|---|---|
| Amaranthin/amaranthin/aerolysin | 0 | 0 | 16 | 0 | 0 |  |
| CRA | 2 | 5 | 4 | 2 | 3 |  |
| CRA/CID | 7 | 0 | 0 | 0 | 0 |  |
| EUL(1–2)* | 1 | 3 | 1 | 5 | 5 |  |
| GNA | 6 | 9 | 3 | 2 | 11 |  |
| GNA/SG(0-2)/PAN(0-2)/PK (0–2)/SRK (0–2) | 43 | 155 | 42 | 125 | 76 |  |
| Hevein(1 or 4) | 0 | 0 | 0 | 2 | 2 |  |
| Hevein/barwin | 1 | 2 | 0 | 0 | 0 |  |
| Hevein(1–2)/GH19 | 9 | 4 | 4 | 8 | 8 |  |
| Jacalin(1–4) | 44 | 4 | 5 | 17 | 21 |  |
| F-box(0–1)/F-box AD1(0–1)/jacalin | 2 | 1 | 0 | 0 | 0 |  |
| Jacalin(1–2)/Kelch1(1,2 or 4) | 4 | 0 | 0 | 0 | 0 |  |
| C1-like(1–3)/DC1/jacalin | 0 | 0 | 3 | 0 | 0 |  |
| PK/jacalin(1–3) | 0 | 0 | 0 | 4 | 1 |  |
| Dirigent/jacalin | 0 | 0 | 0 | 4 | 4 |  |
| NB-ARC/jacalin(1–3) | 0 | 0 | 0 | 3 | 4 |  |
| Legume(1–2) | 13 | 27 | 2 | 18 | 12 |  |
| Legume(1–2)/PK | 41 | 66 | 27 | 79 | 68 |  |
| LysM(1–2) | 6 | 25 | 3 | 12 | 11 |  |
| F-box/LysM | 1 | 2 | 1 | 1 | 0 |  |
| LysM(1–2)/PK | 5 | 18 | 6 | 7 | 10 |  |
| Nictaba(1–2) | 7 | 6 | 8 | 4 | 4 |  |
| F-box/Nictaba(1–2) | 18 | 25 | 12 | 15 | 16 |  |
| TIR/Nictaba | 4 | 0 | 0 | 0 | 0 |  |
| GH5/ricin B | 2 | 6 | 4 | 3 | 3 |  |
| GH27/ricin B | 0 | 4 | 0 | 1 | 1 |  |
| Ricin B/ricin B | 0 | 0 | 3 | 0 | 0 |  |
| RIP/ricin B/ricin B | 0 | 0 | 2 | 0 | 0 |  |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Holle, S.; De Schutter, K.; Eggermont, L.; Tsaneva, M.; Dang, L.; Van Damme, E.J.M. Comparative Study of Lectin Domains in Model Species: New Insights into Evolutionary Dynamics. Int. J. Mol. Sci. 2017, 18, 1136. https://doi.org/10.3390/ijms18061136
Van Holle S, De Schutter K, Eggermont L, Tsaneva M, Dang L, Van Damme EJM. Comparative Study of Lectin Domains in Model Species: New Insights into Evolutionary Dynamics. International Journal of Molecular Sciences. 2017; 18(6):1136. https://doi.org/10.3390/ijms18061136
Chicago/Turabian StyleVan Holle, Sofie, Kristof De Schutter, Lore Eggermont, Mariya Tsaneva, Liuyi Dang, and Els J. M. Van Damme. 2017. "Comparative Study of Lectin Domains in Model Species: New Insights into Evolutionary Dynamics" International Journal of Molecular Sciences 18, no. 6: 1136. https://doi.org/10.3390/ijms18061136








