B-Myb Is Up-Regulated and Promotes Cell Growth and Motility in Non-Small Cell Lung Cancer
Abstract
:1. Introduction
2. Results
2.1. B-Myb Expression Is Up-Regulated in NSCLC
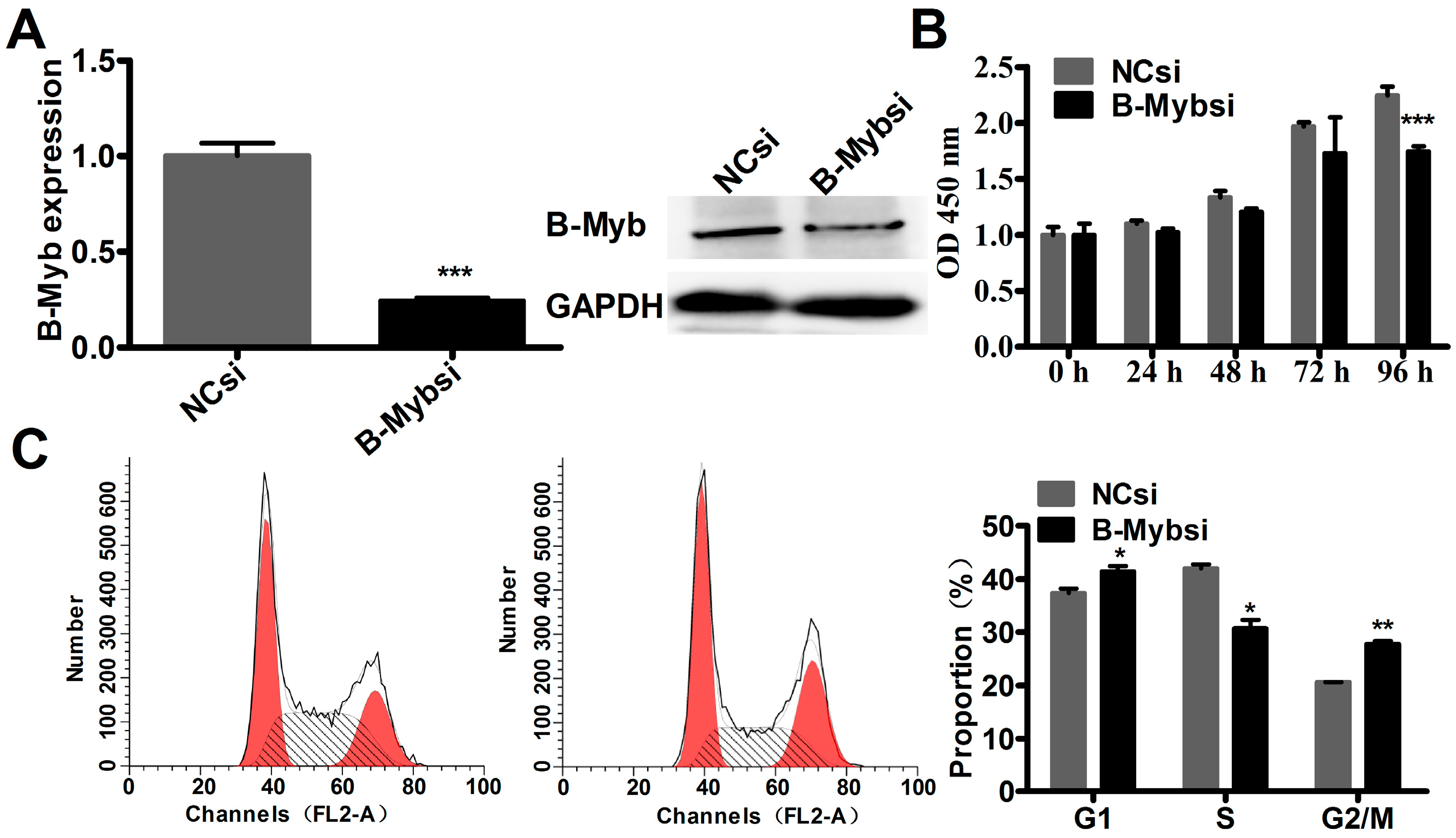
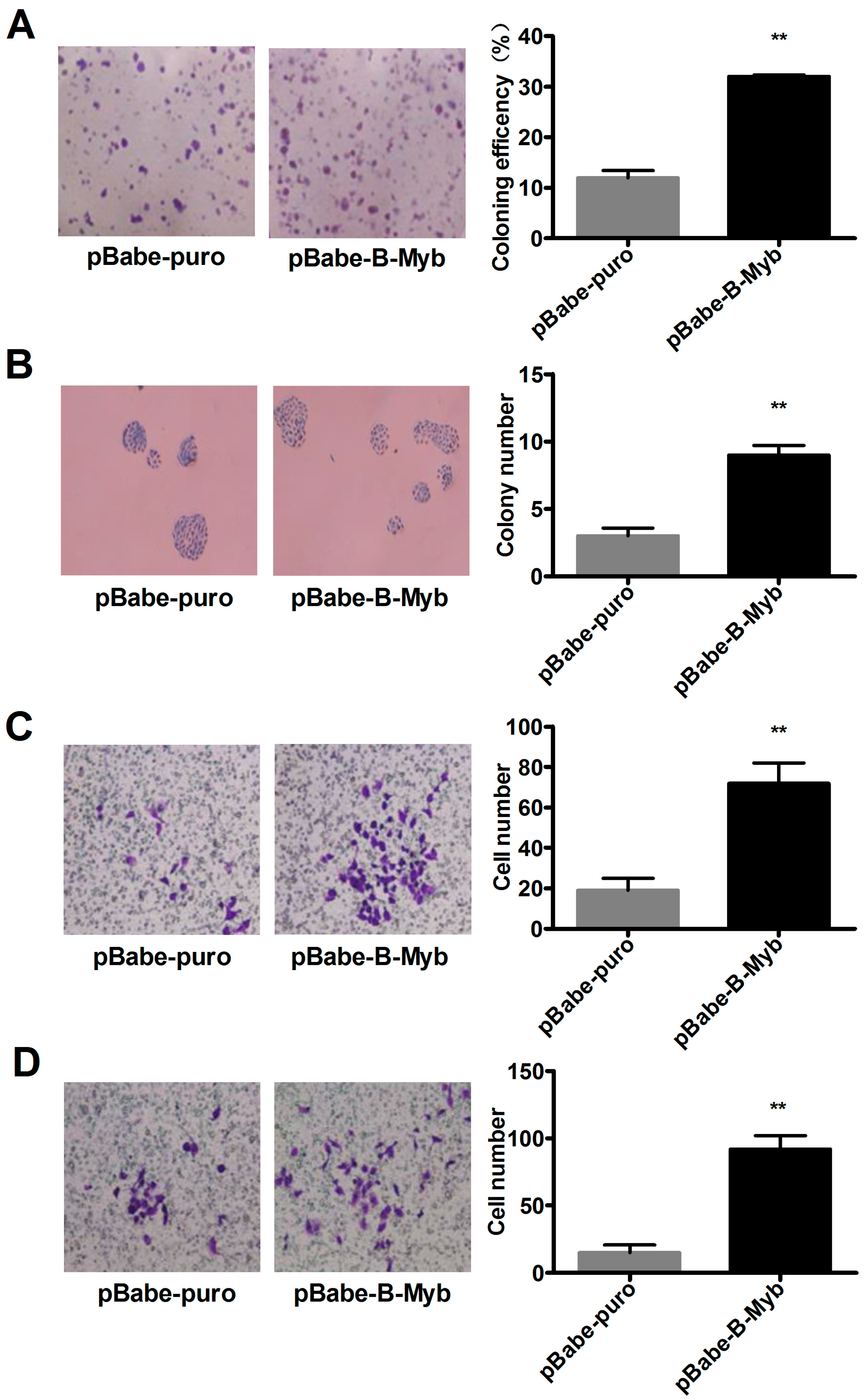
2.2. B-Myb Increases Lung Cancer Cell Growth
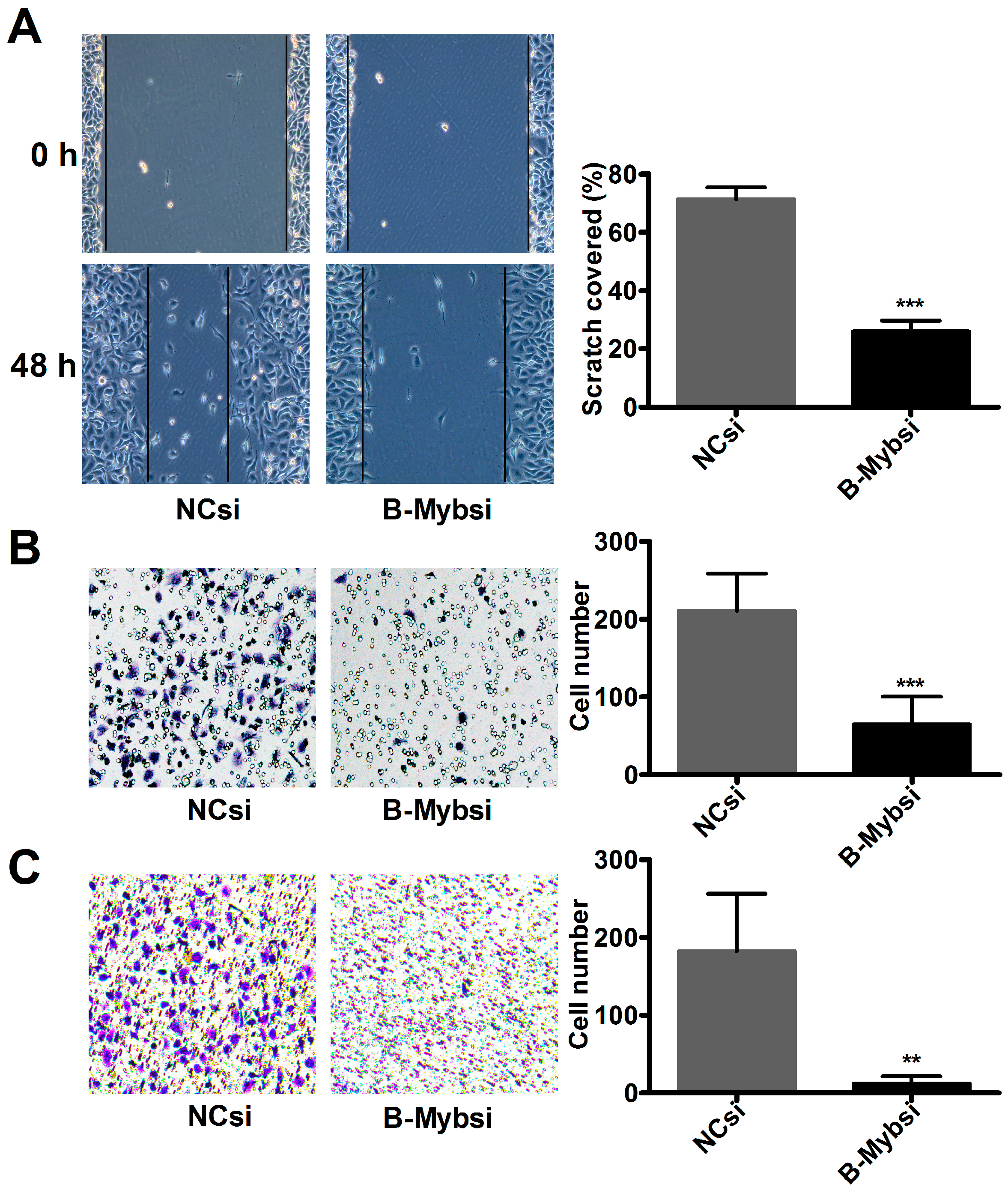
2.3. B-Myb Promotes Lung Cancer Cell Migration and Invasion
2.4. B-Myb Enhances Lung Tumorigenesis In Vivo
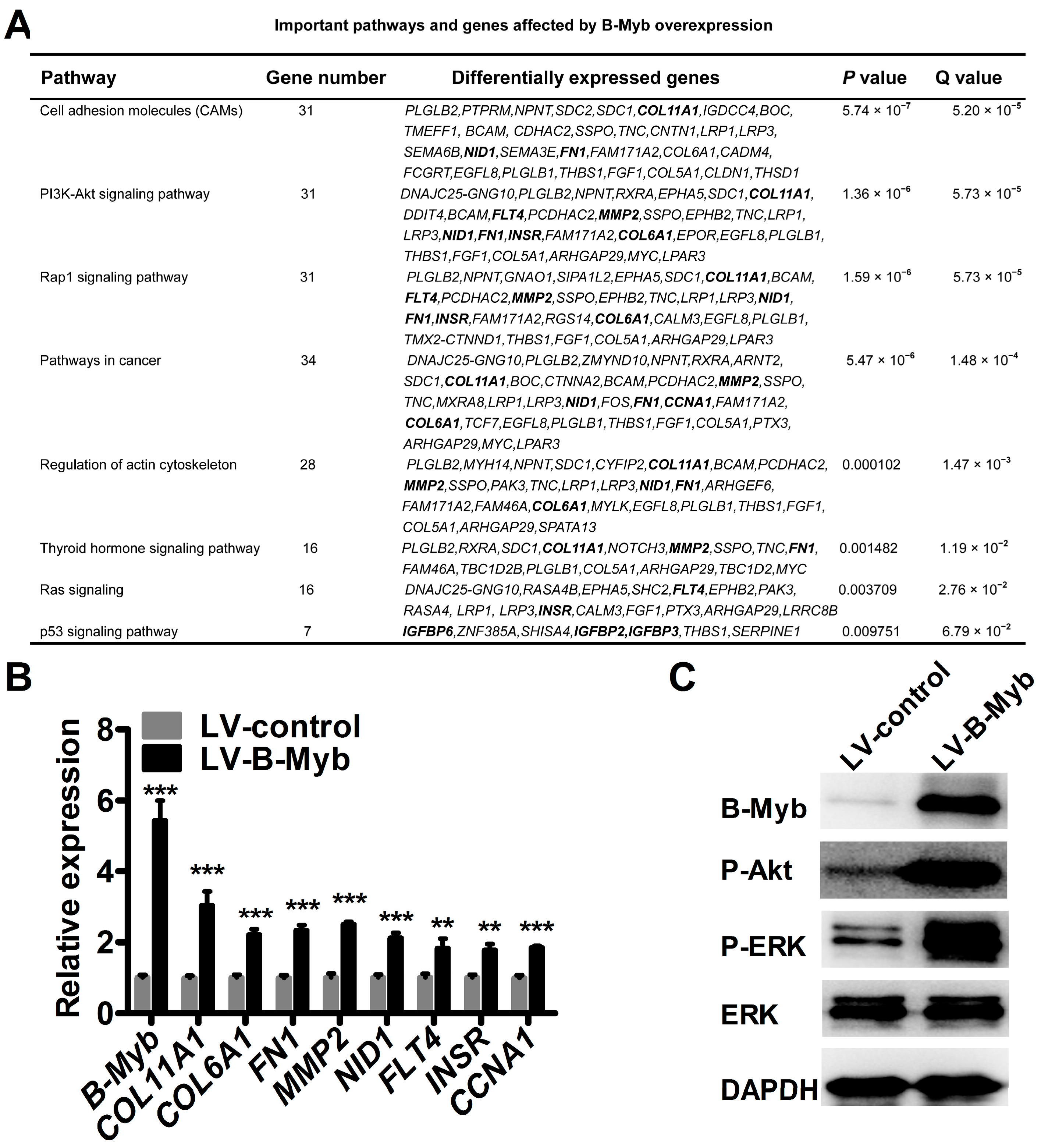
2.5. B-Myb Regulates Various Downstream Genes and Pathways
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. siRNA Synthesis and Transfection
4.3. RNA Isolation and RT-PCR
4.4. Tissue Microarrays and Immunohistochemistry
4.5. pBabe-Mediated Establishment of Stable B-Myb Overexpression Cell Line
4.6. Lentivirus-Mediated Establishment of Stable B-Myb Overexpression Cell Line
4.7. Immunoblot Analysis
4.8. Cell Proliferation Assay
4.9. Cell Cycle Analysis
4.10. Colony Formation Assay
4.11. Wound Healing Assay
4.12. Transwell Cell Migration and Invasion Assays
4.13. RNA-Sequencing Analysis
4.14. Tumor Xenografts
4.15. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NSCLC | non-small cell lung cancer |
| SQCC | lung squamous cell carcinoma |
| ADC | lung adenocarcinoma |
| SCLC | small cell lung cancer |
| Mybl2 | MYB proto-oncogene like 2 |
| COL11A1 | collagen type XI alpha 1 chain |
| COL6A1 | collagen type VI alpha 1 chain |
| FNT | putative formate/nitrite transporter |
| MMP2 | matrix metallopeptidase 2 |
| NID1 | nidogen 1 |
| FLT4 | fms related tyrosine kinase 4 |
| INSR | insulin receptor |
| CCNA1 | cyclin A1 |
| ERK | extracellular regulated MAP kinase |
| Sp1 | Sp1 transcription factor |
| PDK1 | pyruvate dehydrogenase kinase 1 |
References
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- Oh, I.H.; Reddy, E.P. The Myb gene family in cell growth, differentiation and apoptosis. Oncogene 1999, 18, 3017–3033. [Google Scholar] [CrossRef] [PubMed]
- Joaquin, M.; Watson, R.J. Cell cycle regulation by the B-Myb transcription factor. Cell. Mol. Life Sci. 2003, 60, 2389–2401. [Google Scholar] [CrossRef] [PubMed]
- Sala, A. B-Myb, a transcription factor implicated in regulating cell cycle, apoptosis and cancer. Eur. J. Cancer 2005, 41, 2479–2484. [Google Scholar] [CrossRef] [PubMed]
- Martinez, I.; Dimaio, D. B-Myb, cancer, senescence, and microRNAs. Cancer Res. 2011, 71, 5370–5373. [Google Scholar] [CrossRef] [PubMed]
- Mowla, S.N.; Lam, E.W.; Jat, P.S. Cellular senescence and aging: The role of B-Myb. Aging Cell 2014, 13, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Wang, L.; Shen, X.; Xiao, X.; Liu, Z.; Wei, P.; Wang, Y.; Qi, P.; Shen, C.; Sheng, W.; et al. MYBL2 is an independent prognostic marker that has tumor-promoting functions in colorectal cancer. Am. J. Cancer Res. 2015, 5, 1542–1552. [Google Scholar] [PubMed]
- Calvisi, D.F.; Simile, M.M.; Ladu, S.; Frau, M.; Evert, M.; Tomasi, M.L.; Demartis, M.I.; Daino, L.; Seddaiu, M.A.; Brozzetti, S.; et al. Activation of V-Myb avian myeloblastosis viral oncogene homolog-like2 (MYBL2)-LIN9 complex contributes to human hepatocarcinogenesis and identifies a subset of hepatocellular carcinoma with mutant p53. Hepatology 2011, 53, 1226–1236. [Google Scholar] [CrossRef] [PubMed]
- Frau, M.; Ladu, S.; Calvisi, D.F.; Simile, M.M.; Bonelli, P.; Daino, L.; Tomasi, M.L.; Seddaiu, M.A.; Feo, F.; Pascale, R.M. MYBL2 expression is under genetic control and contributes to determine a hepatocellular carcinoma susceptible phenotype. J. Hepatol. 2011, 55, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Yasui, K.; Zen, K.; Inagaki, Y.; Fujii, H.; Minami, M.; Tanaka, S.; Taniwaki, M.; Itoh, Y.; Arii, S.; et al. Activation of B-Myb by E2F1 in hepatocellular carcinoma. Hepatol. Res. 2008, 38, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Ansieau, S.; Kowenz-Leutz, E.; Dechend, R.; Leutz, A. B-Myb, a repressed trans-activating protein. J. Mol. Med. 1997, 75, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Raschella, G.; Negroni, A.; Sala, A.; Pucci, S.; Romeo, A.; Calabretta, B. Requirement of B-Myb function for survival and differentiative potential of human neuroblastoma cells. J. Biol. Chem. 1995, 270, 8540–8545. [Google Scholar] [CrossRef] [PubMed]
- Ahlbory, D.; Appl, H.; Lang, D.; Klempnauer, K.H. Disruption of B-Myb in DT40 cells reveals novel function for B-Myb in the response to DNA-damage. Oncogene 2005, 24, 7127–7134. [Google Scholar] [CrossRef] [PubMed]
- Bar-Shira, A.; Pinthus, J.H.; Rozovsky, U.; Goldstein, M.; Sellers, W.R.; Yaron, Y.; Eshhar, Z.; Orr-Urtreger, A. Multiple genes in human 20q13 chromosomal region are involved in an advanced prostate cancer xenograft. Cancer Res. 2002, 62, 6803–6807. [Google Scholar] [PubMed]
- Bessa, M.; Joaquin, M.; Tavner, F.; Saville, M.K.; Watson, R.J. Regulation of the cell cycle by B-Myb. Blood Cells Mol. Dis. 2001, 27, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Thorner, A.R.; Hoadley, K.A.; Parker, J.S.; Winkel, S.; Millikan, R.C.; Perou, C.M. In vitro and in vivo analysis of B-Myb in basal-like breast cancer. Oncogene 2009, 28, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Long, E.M.; Long, M.A.; Tsirigotis, M.; Gray, D.A. Stimulation of the murine UCHL1 gene promoter by the B-Myb transcription factor. Lung Cancer 2003, 42, 9–21. [Google Scholar] [CrossRef]
- Atjanasuppat, K.; Lirdprapamongkol, K.; Jantaree, P.; Svasti, J. Non-adherent culture induces paclitaxel resistance in H460 lung cancer cells via ERK-mediated up-regulation of betaIVa-tubulin. Biochem. Biophys. Res. Commun. 2015, 466, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, W.C.; Weiss, W.A. Addiction to B-Myb. Oncotarget 2010, 1, 235–236. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Orchard, G.; Lillington, D.M.; Russell-Jones, R.; Young, B.D.; Whittaker, S.J. Amplification and overexpression of JUNB is associated with primary cutaneous T-cell lymphomas. Blood 2003, 101, 1513–1519. [Google Scholar] [CrossRef] [PubMed]
- Fuster, O.; Llop, M.; Dolz, S.; Garcia, P.; Such, E.; Ibanez, M.; Luna, I.; Gomez, I.; Lopez, M.; Cervera, J.; et al. Adverse prognostic value of MYBL2 overexpression and association with microRNA-30 family in acute myeloid leukemia patients. Leuk. Res. 2013, 37, 1690–1696. [Google Scholar] [CrossRef] [PubMed]
- Bartusel, T.; Schubert, S.; Klempnauer, K.H. Regulation of the cyclin D1 and cyclin A1 promoters by B-Myb is mediated by Sp1 binding sites. Gene 2005, 351, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Muller-Tidow, C.; Wang, W.; Idos, G.E.; Diederichs, S.; Yang, R.; Readhead, C.; Berdel, W.E.; Serve, H.; Saville, M.; Watson, R.; et al. Cyclin A1 directly interacts with B-Myb and cyclin A1/cdk2 phosphorylate B-Myb at functionally important serine and threonine residues: Tissue-specific regulation of B-Myb function. Blood 2001, 97, 2091–2097. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.H.; Chang, T.H.; Huang, Y.F.; Chen, C.C.; Chou, C.Y. COL11A1 confers chemoresistance on ovarian cancer cells through the activation of Akt/c/EBPβ pathway and PDK1 stabilization. Oncotarget 2015, 6, 23748–23763. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Yang, M.; Lin, Q.; Zhang, Z.; Zhu, B.; Miao, C. COL11A1 is overexpressed in recurrent non-small cell lung cancer and promotes cell proliferation, migration, invasion and drug resistance. Oncol. Rep. 2016, 36, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.H.; Chang, Y.H.; Wu, Y.S.; Lee, S.H.; Liao, P.C. Quantitative secretome analysis reveals that COL6A1 is a metastasis-associated protein using stacking gel-aided purification combined with iTRAQ labeling. J. Proteome Res. 2011, 10, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Bae, G.Y.; Choi, S.J.; Lee, J.S.; Jo, J.; Lee, J.; Kim, J.; Cha, H.J. Loss of E-cadherin activates EGFR-MEK/ERK signaling, which promotes invasion via the ZEB1/MMP2 axis in non-small cell lung cancer. Oncotarget 2013, 4, 2512–2522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, B.; Liu, Y.; Yao, H.; Lu, N.; Li, B.; Gao, J.; Guo, S.; Han, N.; Qi, J.; et al. The ovarian cancer-derived secretory/releasing proteome: A repertoire of tumor markers. Proteomics 2012, 12, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Chosa, N.; Sawada, S.; Kondo, H.; Yaegashi, T.; Ishisaki, A. VEGF-C and TGF-beta reciprocally regulate mesenchymal stem cell commitment to differentiation into lymphatic endothelial or osteoblastic phenotypes. Int. J. Mol. Med. 2016, 37, 1005–1013. [Google Scholar] [PubMed]
- Zhou, Y.; Zhu, Y.; Fan, X.; Zhang, C.; Wang, Y.; Zhang, L.; Zhang, H.; Wen, T.; Zhang, K.; Huo, X.; et al. NID1, a new regulator of EMT required for metastasis and chemoresistance of ovarian cancer cells. Oncotarget 2017, in press. [Google Scholar] [CrossRef]
- Ji, Y.; Xie, M.; Lan, H.; Zhang, Y.; Long, Y.; Weng, H.; Li, D.; Cai, W.; Zhu, H.; Niu, Y.; et al. PRR11 is a novel gene implicated in cell cycle progression and lung cancer. Int. J. Biochem. Cell Biol. 2013, 45, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Jiang, G.; Zhang, Y.; Li, J.; Li, X.; Yue, J.; Chen, F.; Liu, H.; Li, H.; Zhu, S.; et al. Analysis of transcriptome differences between resistant and susceptible strains of the citrus red mite Panonychus citri (Acari: Tetranychidae). PLoS ONE 2011, 6, e28516. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
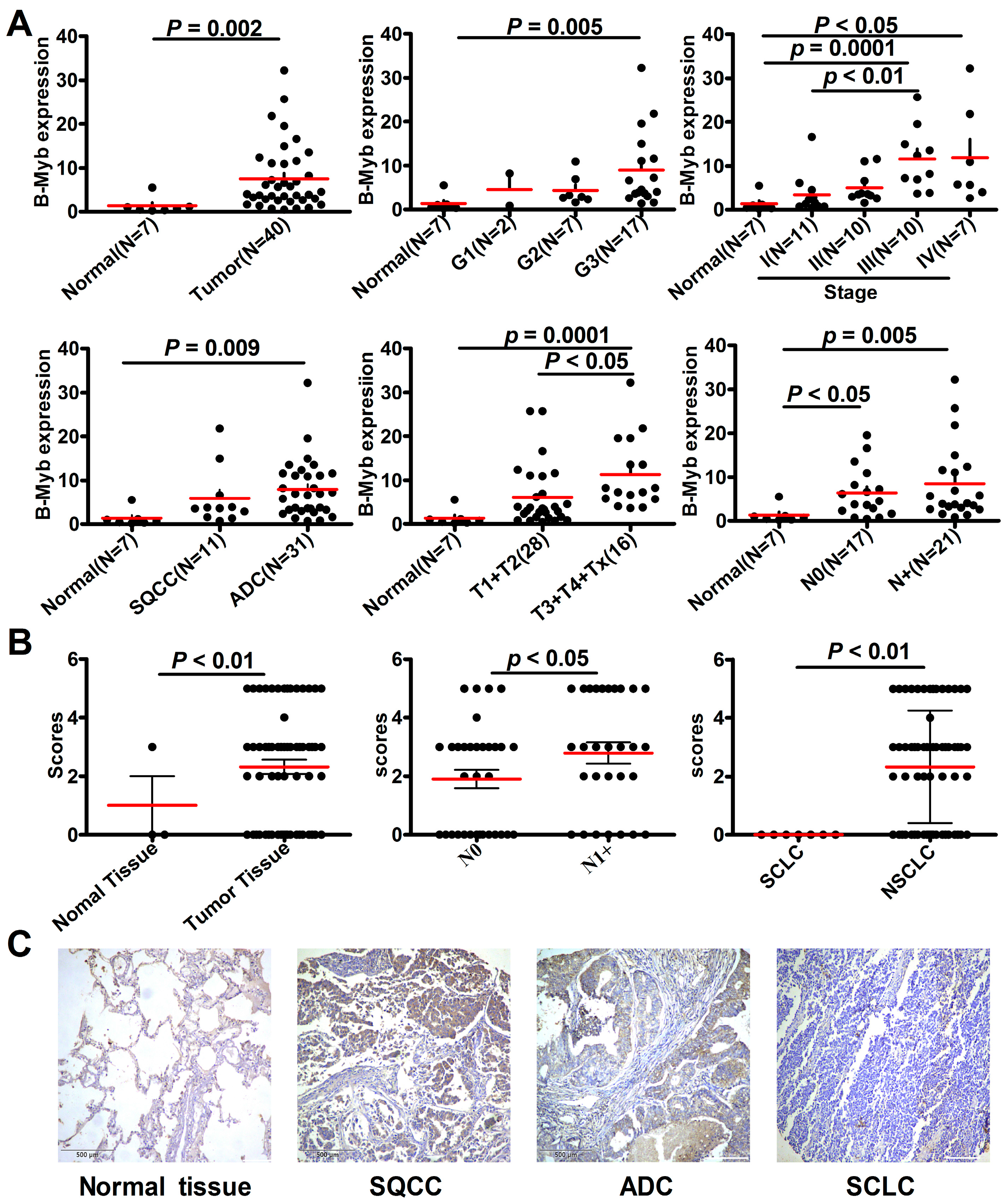


| Pathological Variables | Sample No. | B-Myb IHC Staining (%) | p Value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Normal tissues | 3 | 2 (66.7) | 1 (33.3) | 0.000 *** |
| Lung cancer tissues | 69 | 28 (40.6) | 41 (59.4) | |
| Pathological diagnosis | 0.001 *** | |||
| Non-small cell lung cancer | 62 | 21 (33.9) | 41 (66.1) | |
| Small cell lung cancer | 7 | 7 (100) | 0 (0) | |
| Non-small cell lung cancer | 0.000 *** | |||
| Adenocarcinoma | 28 | 10 (35.7) | 18(64.3) | |
| Atypical carcinoid | 6 | 6 (100) | 0 (0) | |
| Large cell carcinoma | 2 | 1 (50) | 1 (50) | |
| Squamous cell carcinoma | 26 | 4 (15.4) | 22 (84.6) | |
| Sex | 0.408 | |||
| Male | 51 | 19 (37.25) | 32 (62.75) | |
| Female | 18 | 9 (50) | 9 (50) | |
| Stage | 1.000 | |||
| I | 24 | 10 (41.7) | 14 (58.3) | |
| II | 24 | 9 (37.5) | 15 (62.5) | |
| III + IV | 21 | 8 (38.1) | 13 (61.9) | |
| Lymph node status | 0.030 * | |||
| pN0 | 35 | 21 (60.0) | 14 (40.0) | |
| pN1+ | 34 | 11 (32.4) | 23 (67.6) | |
| Tumor size | 0.096 | |||
| T1 | 4 | 3 (75) | 1 (25) | |
| T2 | 43 | 11 (25.6) | 32 (74.4) | |
| T3 | 8 | 3 (37.5) | 5 (62.5) | |
| T4 | 7 | 4 (57.1) | 3 (42.9) | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, Y.; Zhu, H.; Cai, W.; Fan, X.; Wang, Y.; Niu, Y.; Song, F.; Bu, Y. B-Myb Is Up-Regulated and Promotes Cell Growth and Motility in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2017, 18, 860. https://doi.org/10.3390/ijms18060860
Jin Y, Zhu H, Cai W, Fan X, Wang Y, Niu Y, Song F, Bu Y. B-Myb Is Up-Regulated and Promotes Cell Growth and Motility in Non-Small Cell Lung Cancer. International Journal of Molecular Sciences. 2017; 18(6):860. https://doi.org/10.3390/ijms18060860
Chicago/Turabian StyleJin, Yuelei, Huifang Zhu, Wei Cai, Xiaoyan Fan, Yitao Wang, Yulong Niu, Fangzhou Song, and Youquan Bu. 2017. "B-Myb Is Up-Regulated and Promotes Cell Growth and Motility in Non-Small Cell Lung Cancer" International Journal of Molecular Sciences 18, no. 6: 860. https://doi.org/10.3390/ijms18060860





