Associations of Drug Lipophilicity and Extent of Metabolism with Drug-Induced Liver Injury
Abstract
:1. Introduction
2. Results
2.1. Lipophilicity and DILI Risk
2.2. Metabolism and DILI Risk
3. Discussion
4. Materials and Methods
4.1. Drug Datasets
4.2. Daily Dose, Lipophilicity, and Metabolism
4.3. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chalasani, N.; Björnsson, E. Risk factors for idiosyncratic drug-induced liver injury. Gastroenterology 2010, 138, 2246–2259. [Google Scholar] [CrossRef] [PubMed]
- Amacher, D.E. The primary role of hepatic metabolism in idiosyncratic drug-induced liver injury. Expert Opin. Drug Metab. Toxicol. 2012, 8, 335–347. [Google Scholar] [CrossRef] [PubMed]
- Fontana, R.J. Pathogenesis of idiosyncratic drug-induced liver injury and clinical perspectives. Gastroenterology 2014, 146, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Suzuki, A.; Borlak, J.; Andrade, R.J.; Lucena, M.I. Drug-induced liver injury: Interactions between drug properties and host factors. J. Hepatol. 2015, 63, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Daly, A.K.; Donaldson, P.T.; Bhatnagar, P.; Shen, Y.; Pe’er, I.; Floratos, A.; Daly, M.J.; Goldstein, D.B.; John, S.; Nelson, M.R.; et al. HLA-b* 5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat. Genet. 2009, 41, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Lucena, M.I.; Molokhia, M.; Shen, Y.; Urban, T.J.; Aithal, G.P.; Andrade, R.J.; Day, C.P.; Ruiz-Cabello, F.; Donaldson, P.T.; Stephens, C.; et al. Susceptibility to amoxicillin-clavulanate-induced liver injury is influenced by multiple HLA class I and II alleles. Gastroenterology 2011, 141, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Monshi, M.M.; Faulkner, L.; Gibson, A.; Jenkins, R.E.; Farrell, J.; Earnshaw, C.J.; Alfirevic, A.; Cederbrant, K.; Daly, A.K.; French, N.; et al. Human leukocyte antigen (HLA)-B* 57: 01-Restricted activation of drug-specific T cells provides the immunological basis for flucloxacillin-induced liver injury. Hepatology 2013, 57, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, P.T.; Daly, A.K.; Henderson, J.; Graham, J.; Pirmohamed, M.; Bernal, W.; Day, C.P.; Aithal, G.P. Human leucocyte antigen class II genotype in susceptibility and resistance to co-amoxiclav-induced liver injury. J. Hepatol. 2010, 53, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Spraggs, C.F.; Budde, L.R.; Briley, L.P.; Bing, N.; Cox, C.J.; King, K.S.; Whittaker, J.C.; Mooser, V.E.; Preston, A.J.; Stein, S.H.; et al. HLA-DQA1* 02:01 is a major risk factor for lapatinib-induced hepatotoxicity in women with advanced breast cancer. J. Clin. Oncol. 2011, 29, 667–673. [Google Scholar] [CrossRef] [PubMed]
- Leise, M.D.; Poterucha, J.J.; Talwalkar, J.A. Drug-induced liver injury. In Mayo Clinic Proceedings; Elsevier: Amsterdam, the Netherlands, 2014; pp. 95–106. [Google Scholar]
- Lauschke, V.M.; Ingelman-Sundberg, M. The importance of patient-specific factors for hepatic drug response and toxicity. Int. J. Mol. Sci. 2016, 17, 1714. [Google Scholar] [CrossRef] [PubMed]
- Borlak, J.; Chatterji, B.; Londhe, K.B.; Watkins, P.B. Serum acute phase reactants hallmark healthy individuals at risk for acetaminophen-induced liver injury. Genome Med. 2013, 5, 86. [Google Scholar] [CrossRef] [PubMed]
- Lammert, C.; Einarsson, S.; Saha, C.; Niklasson, A.; Bjornsson, E.; Chalasani, N. Relationship between daily dose of oral medications and idiosyncratic drug-induced liver injury: Search for signals. Hepatology 2008, 47, 2003–2009. [Google Scholar] [CrossRef] [PubMed]
- Lucena, M.I.; Andrade, R.J.; Kaplowitz, N.; García-Cortes, M.; Fernández, M.C.; Romero-Gomez, M.; Bruguera, M.; Hallal, H.; Robles-Diaz, M.; Rodriguez-González, J.F.; et al. Phenotypic characterization of idiosyncratic drug-induced liver injury: The influence of age and sex. Hepatology 2009, 49, 2001–2009. [Google Scholar] [CrossRef] [PubMed]
- Lammert, C.; Bjornsson, E.; Niklasson, A.; Chalasani, N. Oral medications with significant hepatic metabolism at higher risk for hepatic adverse events. Hepatology 2010, 51, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Borlak, J.; Tong, W. High lipophilicity and high daily dose of oral medications are associated with significant risk for drug-induced liver injury. Hepatology 2013, 58, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Suzuki, A.; Thakkar, S.; Yu, K.; Hu, C.; Tong, W. Dilirank: The largest reference drug list ranked by the risk for developing drug-induced liver injury in humans. Drug Discov. Today 2016, 21, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. Venndiagram: A package for the generation of highly-customizable Venn and Euler diagrams in r. BMC Bioinform. 2011, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Greene, N.; Fisk, L.; Naven, R.T.; Note, R.R.; Patel, M.L.; Pelletier, D.J. Developing structure—Activity relationships for the prediction of hepatotoxicity. Chem. Res. Toxicol. 2010, 23, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Kruhlak, N.L. Construction and analysis of a human hepatotoxicity database suitable for QSAR modeling using post-market safety data. Toxicology 2014, 321, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Sakatis, M.Z.; Reese, M.J.; Harrell, A.W.; Taylor, M.A.; Baines, I.A.; Chen, L.; Bloomer, J.C.; Yang, E.Y.; Ellens, H.M.; Ambroso, J.L.; et al. Preclinical strategy to reduce clinical hepatotoxicity using in vitro bioactivation data for >200 compounds. Chem. Res. Toxicol. 2012, 25, 2067–2082. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.J.; Henstock, P.V.; Dunn, M.C.; Smith, A.R.; Chabot, J.R.; de Graaf, D. Cellular imaging predictions of clinical drug-induced liver injury. Toxicol. Sci. 2008, 105, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Kaplowitz, N. Avoiding idiosyncratic DILI: Two is better than one. Hepatology 2013, 58, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Danan, G.; Teschke, R. RUCAM in drug and herb induced liver injury: The update. Int. J. Mol. Sci. 2015, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.H. Causality assessment: Which is best—Expert opinion or rucam? Clin. Liver Dis. 2014, 4, 4–8. [Google Scholar] [CrossRef]
- García-Cortés, M.; Lucena, M.; Pachkoria, K.; Borraz, Y.; Hidalgo, R.; Andrade, R. Evaluation of naranjo adverse drug reactions probability scale in causality assessment of drug-induced liver injury. Aliment. Pharmacol. Ther. 2008, 27, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Björnsson, E.S. Hepatotoxicity by drugs: The most common implicated agents. Int. J. Mol. Sci. 2016, 17, 224. [Google Scholar] [CrossRef]
- Teschke, R.; Frenzel, C.; Wolff, A.; Eickhoff, A.; Schulze, J. Drug induced liver injury: Accuracy of diagnosis in published reports. Ann. Hepatol. 2014, 13, 248–255. [Google Scholar] [PubMed]
- Weng, Z.; Wang, K.; Li, H.; Shi, Q. A comprehensive study of the association between drug hepatotoxicity and daily dose, liver metabolism, and lipophilicity using 975 oral medications. Oncotarget 2015, 6, 17031–17038. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Alonso, A.; Stephens, C.; Lucena, M.I.; Andrade, R.J. Case characterization, clinical features and risk factors in drug-induced liver injury. Int. J. Mol. Sci. 2016, 17, 714. [Google Scholar] [CrossRef] [PubMed]
- Corsini, A.; Bortolini, M. Drug-induced liver injury: The role of drug metabolism and transport. J. Clin. Pharmacol. 2013, 53, 463–474. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Andrade, R.J.; Bjornsson, E.; Lucena, M.I.; Lee, W.M.; Yuen, N.A.; Hunt, C.M.; Freston, J.W. Drugs associated with hepatotoxicity and their reporting frequency of liver adverse events in vigibase™. Drug Saf. 2010, 33, 503–522. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. Drugbank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
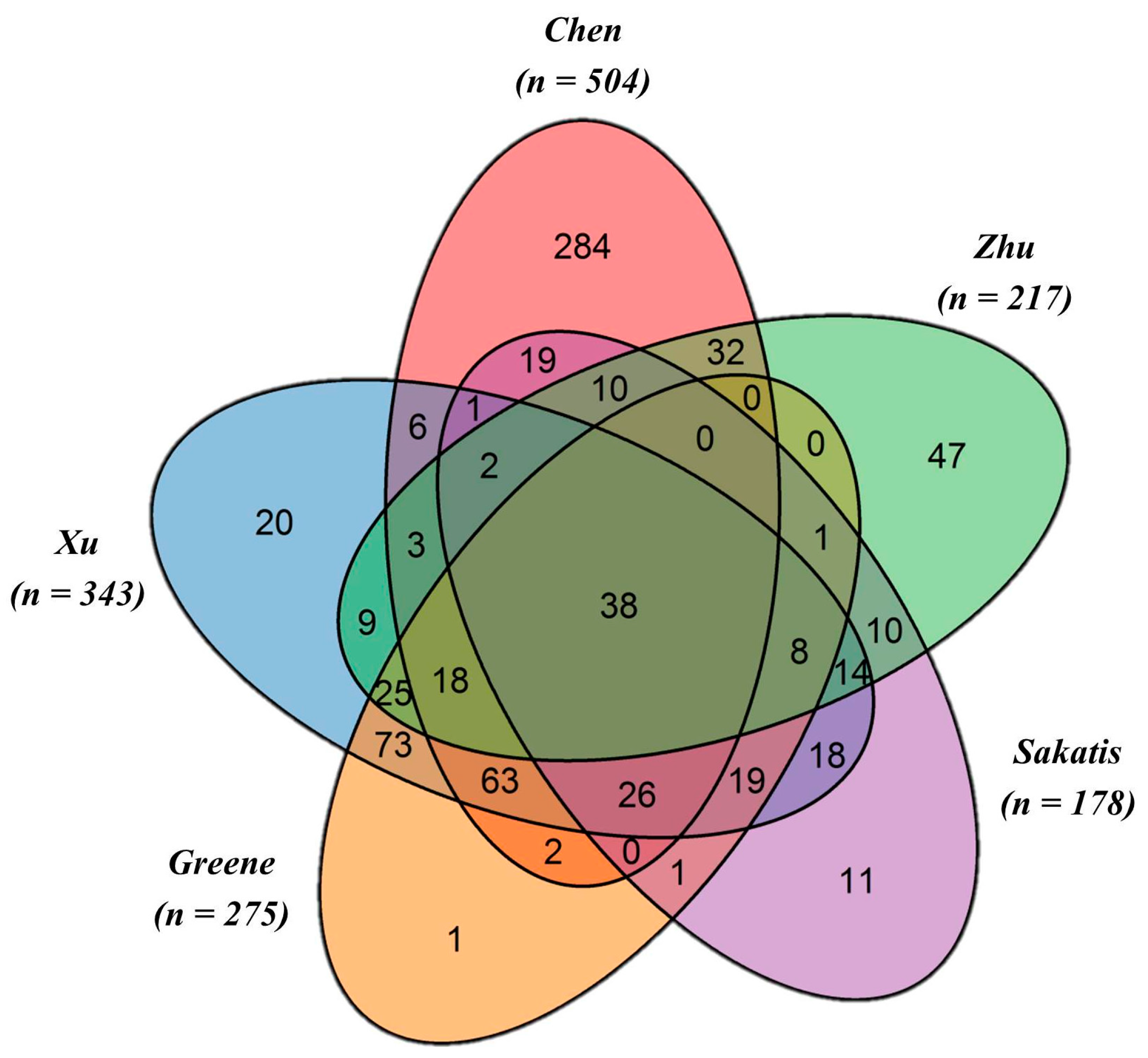
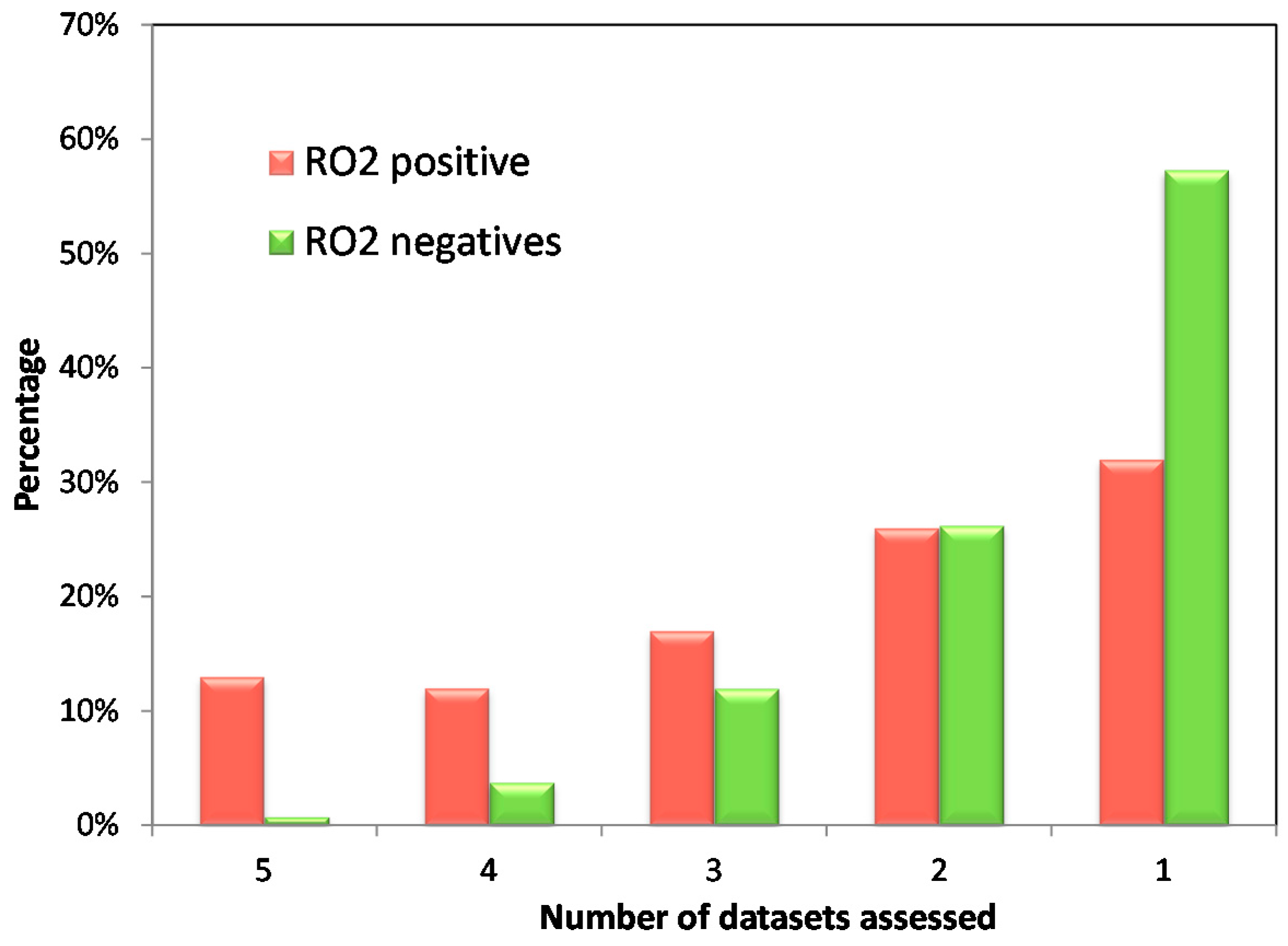
| Annotated Datasets | DILI Classification | Positives | Negatives | Odds Ratio (95% Confidence Interval) | p-Value |
|---|---|---|---|---|---|
| LogP ≥ 3 and Daily Dose ≥ 100 mg | |||||
| Chen [17] | vMost-concern (n = 172) | 71 | 101 | 11.50 (5.42–24.82) | <0.05 |
| vNo-concern (n = 173) | 10 | 163 | |||
| Greene [19] | Human hepatotoxicity (n = 174) | 51 | 123 | 4.08 (1.57–11.23) | <0.05 |
| No evidence (n = 65) | 6 | 59 | |||
| Zhu [20] | Hepatotoxic (n = 152) | 42 | 110 | 5.47 (1.52–23.42) | <0.05 |
| Non-hepatotoxic (n = 46) | 3 | 43 | |||
| Sakatis [21] | Hepatotoxic (n = 89) | 28 | 61 | 2.80 (1.20–6.57) | <0.05 |
| Non-hepatotoxic (n = 78) | 11 | 67 | |||
| Xu [22] | Positive (n = 179) | 56 | 123 | 2.32 (1.27–4.24) | <0.05 |
| Negative (n = 128) | 21 | 107 | |||
| Consensus | Positive (n = 313) | 99 | 214 | 4.77 (2.79–7.86) | <0.05 |
| Negative (n = 255) | 23 | 232 | |||
| LogP ≥ 3 | |||||
| Chen [17] | vMost-concern (n = 172) | 87 | 85 | 2.26 (1.42–3.58) | <0.05 |
| vNo-concern (n = 173) | 54 | 119 | |||
| Greene [19] | Human hepatotoxicity (n = 174) | 69 | 105 | 1.72 (0.88–3.36) | 0.098 |
| No evidence (n = 65) | 18 | 47 | |||
| Zhu [20] | Hepatotoxic (n = 152) | 64 | 88 | 2.31 (1.03–5.26) | <0.05 |
| Non-hepatotoxic (n = 46) | 11 | 35 | |||
| Sakatis [21] | Hepatotoxic (n = 89) | 35 | 54 | 1.03 (0.53–2.03) | 1.0 |
| Non-hepatotoxic (n = 78) | 30 | 48 | |||
| Xu [22] | Positive (n = 179) | 74 | 105 | 0.97 (0.59–1.57) | 0.91 |
| Negative (n = 128) | 54 | 74 | |||
| Consensus | Positive (n = 313) | 138 | 175 | 1.55 (1.09–2.22) | <0.05 |
| Negative (n = 255) | 86 | 169 | |||
| Daily Dose ≥ 100 mg | |||||
| Chen [17] | vMost-concern (n = 172) | 139 | 33 | 6.20 (3.71–10.39) | <0.05 |
| vNo-concern (n = 173) | 70 | 103 | |||
| Greene [19] | Human hepatotoxicity (n = 174) | 123 | 51 | 2.65 (1.41–4.96) | <0.05 |
| No evidence (n = 65) | 31 | 34 | |||
| Zhu [20] | Hepatotoxic (n = 152) | 110 | 42 | 2.86 (1.37–5.96) | <0.05 |
| Non-hepatotoxic (n = 46) | 22 | 24 | |||
| Sakatis [21] | Hepatotoxic (n = 89) | 73 | 16 | 7.3 (3.41–15.83) | <0.05 |
| Non-hepatotoxic (n = 78) | 30 | 48 | |||
| Xu [22] | Positive (n = 179) | 127 | 52 | 2.52 (1.52–4.16) | <0.05 |
| Negative (n = 128) | 63 | 65 | |||
| Consensus | Positive (n = 313) | 225 | 88 | 3.54 (2.46–5.10) | <0.05 |
| Negative (n = 255) | 107 | 148 | |||
| Annotated Datasets | DILI Classification | Positives | Negatives | Odds Ratio (95% Confidence Interval) | p-Value |
|---|---|---|---|---|---|
| Hepatic Metabolism ≥ 50% and Daily Dose ≥ 100 mg | |||||
| Chen [17] | vMost-concern (n = 107) | 76 | 31 | 11.09 (5.54–22.48) | <0.05 |
| vNo-concern (n = 105) | 19 | 86 | |||
| Greene [19] | Human hepatotoxicity (n = 139) | 81 | 58 | 4.32 (1.91–9.92) | <0.05 |
| No evidence (n = 45) | 11 | 34 | |||
| Zhu [20] | Hepatotoxic (n = 127) | 70 | 57 | 5.32 (1.90–15.57) | <0.05 |
| Non-hepatotoxic (n = 32) | 6 | 26 | |||
| Sakatis [21] | Hepatotoxic (n = 73) | 53 | 20 | 7.48 (3.30–17.20) | <0.05 |
| Non-hepatotoxic (n = 65) | 17 | 48 | |||
| Xu [22] | Positive (n = 141) | 84 | 57 | 3.79 (2.11–6.84) | <0.05 |
| Negative (n = 100) | 28 | 72 | |||
| Consensus | Positive (n = 221) | 127 | 94 | 5.48 (3.40–8.84) | <0.05 |
| Negative (n = 177) | 35 | 142 | |||
| Hepatic Metabolism ≥ 50% | |||||
| Chen [17] | vMost-concern (n = 107) | 91 | 16 | 2.67 (1.27–5.40) | <0.05 |
| vNo-concern (n = 105) | 72 | 33 | |||
| Greene [19] | Human hepatotoxicity (n = 139) | 109 | 30 | 2.42 (1.11–5.29) | <0.05 |
| No evidence (n = 45) | 27 | 18 | |||
| Zhu [20] | Hepatotoxic (n = 127) | 98 | 29 | 1.77 (0.70–4.42) | 0.18 |
| Non-hepatotoxic (n = 32) | 21 | 11 | |||
| Sakatis [21] | Hepatotoxic (n = 73) | 61 | 12 | 1.80 (0.73–4.48) | 0.21 |
| Non-hepatotoxic (n = 65) | 48 | 17 | |||
| Xu [22] | Positive (n = 141) | 112 | 29 | 1.92 (1.02–3.56) | <0.05 |
| Negative (n = 100) | 67 | 33 | |||
| Consensus | Positive (n = 221) | 174 | 47 | 1.90 (1.18–3.05) | <0.05 |
| Negative (n = 177) | 117 | 60 | |||
| Daily Dose ≥ 100 mg | |||||
| Chen [17] | vMost-concern (n = 107) | 87 | 20 | 7.67 (3.92–15.15) | <0.05 |
| vNo-concern (n = 105) | 38 | 67 | |||
| Greene [23] | Human hepatotoxicity (n = 139) | 99 | 40 | 2.37 (1.12–5.00) | <0.05 |
| No evidence (n = 45) | 23 | 22 | |||
| Zhu [20] | Hepatotoxic (n = 127) | 90 | 37 | 3.13 (1.32–7.49) | <0.05 |
| Non-hepatotoxic (n = 32) | 14 | 18 | |||
| Sakatis [21] | Hepatotoxic (n = 73) | 61 | 12 | 7.63 (3.23–18.33) | <0.05 |
| Non-hepatotoxic (n = 65) | 26 | 39 | |||
| Xu [22] | Positive (n = 141) | 101 | 40 | 2.43 (1.37–4.30) | <0.05 |
| Negative (n = 100) | 51 | 49 | |||
| Consensus | Positive (n = 221) | 160 | 61 | 4.01 (2.57–6.26) | <0.05 |
| Negative (n = 177) | 70 | 107 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McEuen, K.; Borlak, J.; Tong, W.; Chen, M. Associations of Drug Lipophilicity and Extent of Metabolism with Drug-Induced Liver Injury. Int. J. Mol. Sci. 2017, 18, 1335. https://doi.org/10.3390/ijms18071335
McEuen K, Borlak J, Tong W, Chen M. Associations of Drug Lipophilicity and Extent of Metabolism with Drug-Induced Liver Injury. International Journal of Molecular Sciences. 2017; 18(7):1335. https://doi.org/10.3390/ijms18071335
Chicago/Turabian StyleMcEuen, Kristin, Jürgen Borlak, Weida Tong, and Minjun Chen. 2017. "Associations of Drug Lipophilicity and Extent of Metabolism with Drug-Induced Liver Injury" International Journal of Molecular Sciences 18, no. 7: 1335. https://doi.org/10.3390/ijms18071335





