Integrity and Quantity of Total Cell-Free DNA in the Diagnosis of Thyroid Cancer: Correlation with Cytological Classification
Abstract
:1. Introduction
2. Results
2.1. Plasma DNA Concentration and Integrity in Basal Blood Samples
2.2. cfDNA Fragments
2.3. Plasma DNA Integrity in Post-Surgery Blood Samples
2.4. ROC (Receiver Operating Characteristic) Curve Analysis
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. DNA Extraction
4.3. Plasma DNA Integrity Index 180/67 by qPCR
4.4. Statistical Analysis
Abbreviations
| qPCR | quantitative real-time PCR |
| cfDNA | cell-free DNA |
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fleischhacker, M.; Schmidt, B. Circulating nucleic acids (CNAs) and cancer—A survey. Biochim. Biophys. Acta 2007, 1775, 181–232. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Fleischhacker, M.; Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker—A critical appraisal of the literature. Clin. Chim. Acta 2010, 411, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Van der Vaart, M.; Pretorius, P.J. Circulating DNA— Its origin and fluctuation. Ann. N. Y. Acad. Sci. 2008, 1137, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, P.; Salvianti, F.; Pazzagli, M.; Orlando, C. Circulating nucleic acids in cancer and pregnancy. Methods 2010, 50, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Lo, Y.M. The long and short of circulating cell-free DNA and the Ins and Outs of molecular diagnostics. Trends Genet. 2016, 32, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Umetani, N.; Giuliano, A.E.; Hiramatsu, S.H.; Amersi, F.; Nakagawa, T.; Martino, S.; Hoon, D.S. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J. Clin. Oncol. 2006, 24, 4270–4276. [Google Scholar] [CrossRef] [PubMed]
- Umetani, N.; Kim, J.; Hiramatsu, S.; Reber, H.A.; Hines, O.J.; Bilchik, A.J.; Hoon, D.S. Increased integrity of free circulating DNA in sera of patients with colorectal or periampullary cancer: Direct quantitative PCR for ALU repeats. Clin. Chem. 2006, 52, 1062–1069. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.; Weickmann, S.; Witt, C.; Fleischhacker, M. Integrity of cell-free plasma DNA in patients with lung cancer and nonmalignant lung disease. Ann. N. Y. Acad. Sci. 2008, 1137, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Pinzani, P.; Salvianti, F.; Zaccara, S.; Massi, D.; De Giorgi, V.; Pazzagli, M.; Orlando, C. Circulating cell-free DNA in plasma of melanoma patients: Qualitative and quantitative considerations. Clin. Chim. Acta 2011, 412, 2141–2145. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.G.; Huang, H.Y.; Chen, Y.C.; Bristow, R.E.; Kassauei, K.; Cheng, C.C.; Roden, R.; Sokoll, L.J.; Chan, D.W.; Shih, I. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003, 63, 3966–3968. [Google Scholar] [PubMed]
- Jiang, W.W.; Zahurak, M.; Goldenberg, D.; Milman, Y.; Park, H.L.; Westra, W.H.; Koch, W.; Sidransky, D.; Califano, J. Increased plasma DNA integrity index in head and neck cancer patients. Int. J. Cancer 2006, 119, 2673–2676. [Google Scholar] [CrossRef] [PubMed]
- Hanley, R.; Rieger-Christ, K.M.; Canes, D.; Emara, N.R.; Shuber, A.P.; Boynton, K.A.; Libertino, J.A.; Summerhayes, I.C. DNA integrity assay: A plasma-based screening tool for the detection of prostate cancer. Clin. Cancer Res. 2006, 12, 4569–4574. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Vishnubhatla, S.; Raina, V.; Sharma, S.; Gogia, A.; Deo, S.S.; Mathur, S.; Shukla, N.K. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. Springerplus 2015, 4, 265. [Google Scholar] [CrossRef] [PubMed]
- Kamel, A.M.; Teama, S.; Fawzy, A.; El Deftar, M. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumour Biol. 2016, 37, 7565–7572. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, A.; Sweify, K.M.; El-Fayoumy, H.M.; Nofal, N. Quantitative analysis of plasma cell-free DNA and its DNA integrity in patients with metastatic prostate cancer using ALU sequence. J. Egypt Natl. Canc. Inst. 2016, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Leszinski, G.; Lehner, J.; Gezer, U.; Holdenrieder, S. Increased DNA integrity in colorectal cancer. In Vivo 2014, 28, 299–303. [Google Scholar] [PubMed]
- El-Gayar, D.; El-Abd, N.; Hassan, N.; Ali, R. Increased Free Circulating DNA Integrity Index as a Serum Biomarker in Patients with Colorectal Carcinoma. Asian Pac. J. Cancer Prev. 2016, 17, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Bedin, C.; Enzo, M.V.; Del Bianco, P.; Pucciarelli, S.; Nitti, D.; Agostini, M. Diagnostic and prognostic role of cell-free DNA testing for colorectal cancer patients. Int. J. Cancer 2017, 140, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Szpechcinski, A.; Rudzinski, P.; Kupis, W.; Langfort, R.; Orlowski, T.; Chorostowska-Wynimko, J. Plasma cell-free DNA levels and integrity in patients with chest radiological findings: NSCLC versus benign lung nodules. Cancer Lett. 2016, 374, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Holdenrieder, S.; Burges, A.; Reich, O.; Spelsberg, F.W.; Stieber, P. DNA integrity in plasma and serum of patients with malignant and benign diseases. Ann. N. Y. Acad Sci. 2008, 1137, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Salvianti, F.; Pinzani, P.; Verderio, P.; Ciniselli, C.M.; Massi, D.; De Giorgi, V.; Grazzini, M.; Pazzagli, M.; Orlando, C. Multiparametric analysis of cell-free DNA in melanoma patients. PLoS ONE 2012, 7, e49843. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar] [PubMed]
- Suzuki, N.; Kamataki, A.; Yamaki, J.; Homma, Y. Characterization of circulating DNA in healthy human plasma. Clin. Chim. Acta 2008, 387, 55. [Google Scholar] [CrossRef] [PubMed]
- Gang, F.; Guorong, L.; An, Z.; Anne, G.P.; Christian, G.; Jacques, T. Prediction of clear cell renal cell carcinoma by integrity of cell-free DNA in serum. Urology 2010, 75, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Pupilli, C.; Pinzani, P.; Salvianti, F.; Fibbi, B.; Rossi, M.; Petrone, L.; Perigli, G.; de Feo, M.L.; Vezzosi, V.; Pazzagli, M.; et al. Circulating BRAFV600E in the diagnosis and follow-up of differentiated papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2013, 98, 3359–3365. [Google Scholar] [CrossRef] [PubMed]
- Gharib, H.; Papini, E.; Paschke, R.; Duick, D.S.; Valcavi, R.; Hegedüs, L.; Vitti, P.; AACE/AME/ETA Task Force on Thyroid Nodules. American Association of Clinical Endocrinologists, Associazione Medici Endocrinologi, and European Thyroid Association Medical Guidelines for Clinical Practice for the Diagnosis and Management of Thyroid Nodules. Endocr. Pract. 2010, 16 (Suppl. 1), 1–43. [Google Scholar] [CrossRef] [PubMed]
- British Thyroid Association; Royal College of Physicians. Diagnostic Categories. Guidelines for the Management of Thyroid Cancer, 2nd ed.; Lavenham Press: Suffolk, UK, 2007; p. 10. [Google Scholar]
- Cooper, D.S.; Doherty, G.M.; Haugen, B.R.; Kloos, R.T.; Lee, S.L.; Mandel, S.J.; Mazzaferri, E.L.; McIver, B.; Pacini, F.; Schlumberger, M.; et al. Revised American Thyroid Association management guidelines for patients with thyroid nodules and differentiated thyroid cancer. Thyroid 2009, 19, 1167–1214. [Google Scholar] [CrossRef] [PubMed]
- Pacini, F.; Castagna, M.G.; Brilli, L.; Pentheroudakis, G.; ESMO Guidelines Working Group. Thyroid cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2012, 23 (Suppl. 7), VII110–VII119. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, U.; Glöckner, S.; Kleeberger, W.; von Wasielewski, H.F.; Kreipe, H. Detection of gene amplification in archival breast cancer specimens by laser-assisted microdissection and quantitative real-time polymerase chain reaction. Am. J. Pathol. 2000, 156, 1855–1864. [Google Scholar] [CrossRef]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed]
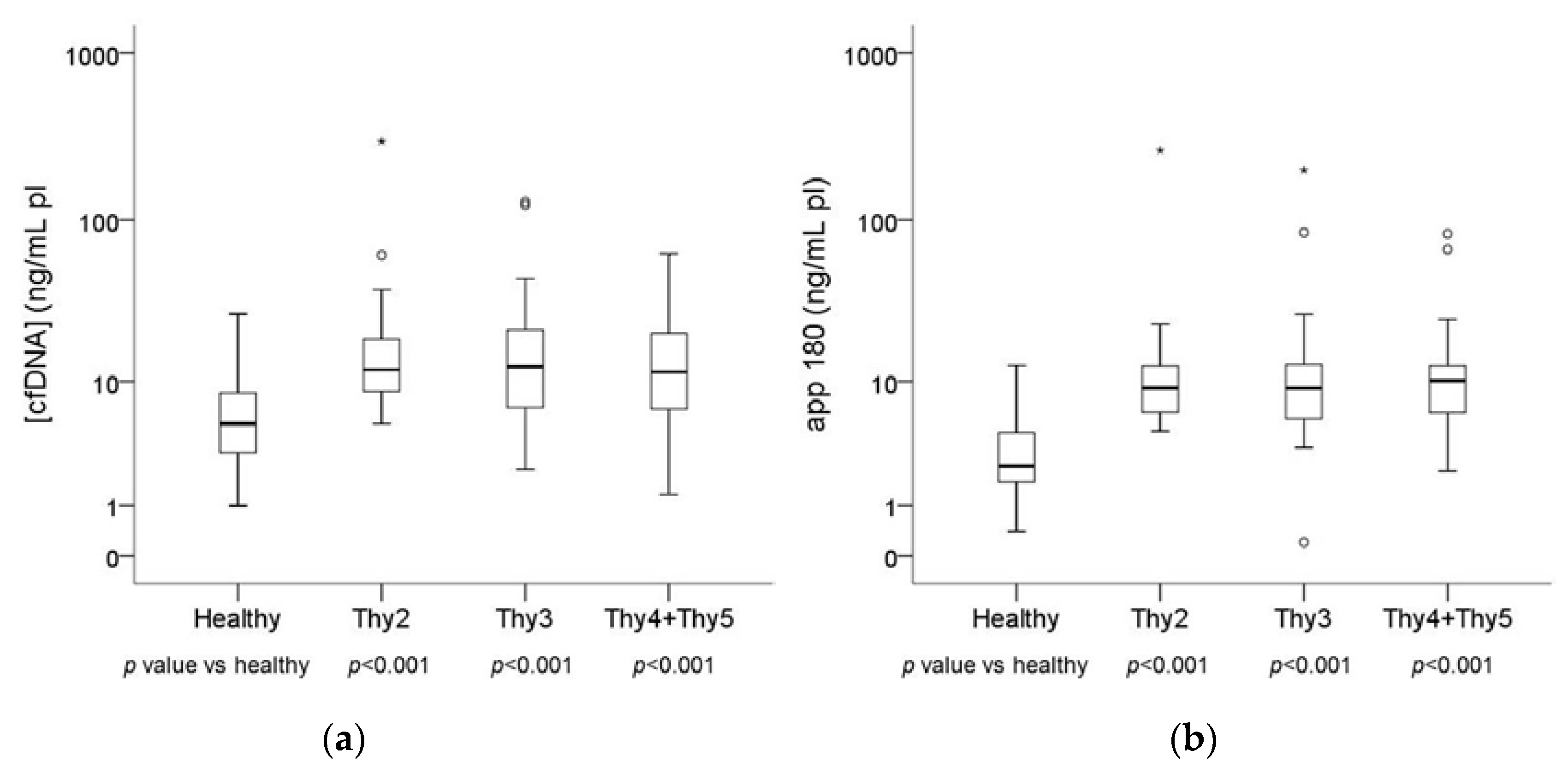
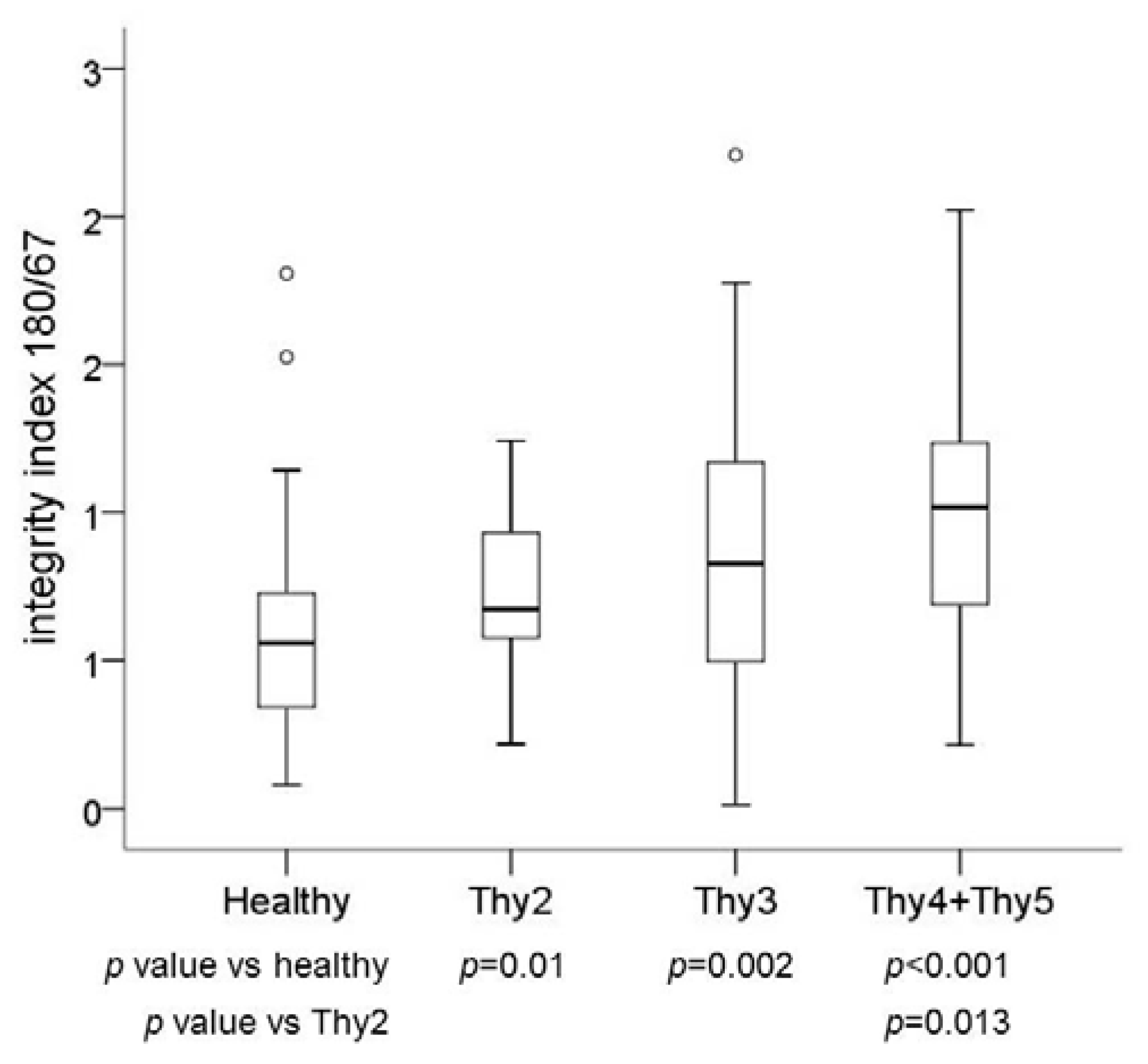

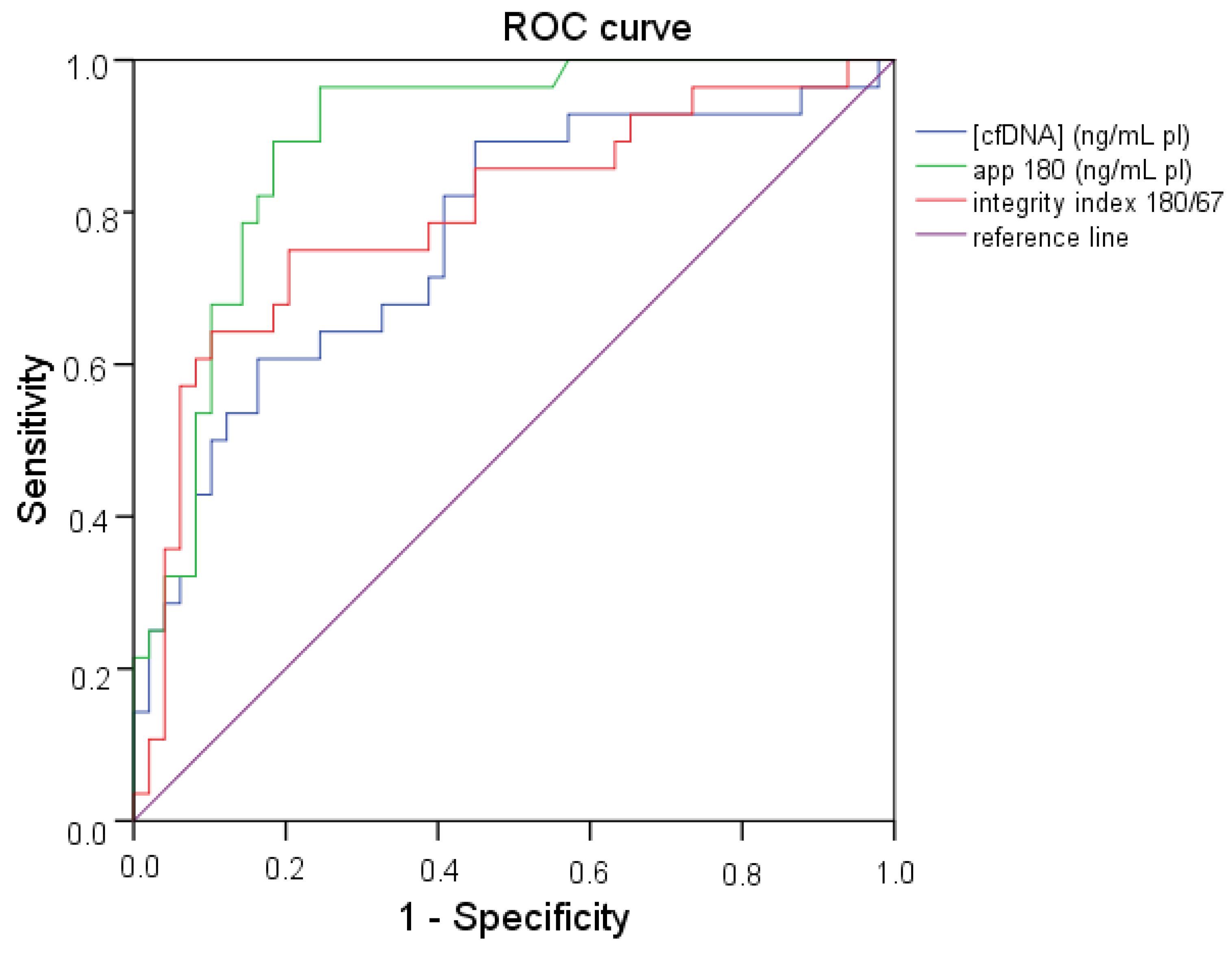
| (cfDNA) (ng/mL pl) | App 180 (ng/mL pl) | Integrity Index 180/67 | ||
|---|---|---|---|---|
| Healthy n = 49 | Median | 5.12 | 2.42 | 0.56 |
| Range | 0.99–26.71 | 0.40–12.68 | 0.08–1.81 | |
| Thy2 n = 25 | Median | 11.88 | 9.03 | 0.67 |
| Range | 5.10–296.52 | 4.55–261.04 | 0.22–1.24 | |
| Thy3 n = 44 | Median | 12.41 | 9.02 | 0.83 |
| Range | 2.26–128.44 | 0.20–199.07 | 0.01–2.21 | |
| Thy4 + Thy5 n = 28 | Median | 11.47 | 10.15 | 1.02 |
| Range | 1.31–62.60 | 2.19–82.65 | 0.22–2.02 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvianti, F.; Giuliani, C.; Petrone, L.; Mancini, I.; Vezzosi, V.; Pupilli, C.; Pinzani, P. Integrity and Quantity of Total Cell-Free DNA in the Diagnosis of Thyroid Cancer: Correlation with Cytological Classification. Int. J. Mol. Sci. 2017, 18, 1350. https://doi.org/10.3390/ijms18071350
Salvianti F, Giuliani C, Petrone L, Mancini I, Vezzosi V, Pupilli C, Pinzani P. Integrity and Quantity of Total Cell-Free DNA in the Diagnosis of Thyroid Cancer: Correlation with Cytological Classification. International Journal of Molecular Sciences. 2017; 18(7):1350. https://doi.org/10.3390/ijms18071350
Chicago/Turabian StyleSalvianti, Francesca, Corinna Giuliani, Luisa Petrone, Irene Mancini, Vania Vezzosi, Cinzia Pupilli, and Pamela Pinzani. 2017. "Integrity and Quantity of Total Cell-Free DNA in the Diagnosis of Thyroid Cancer: Correlation with Cytological Classification" International Journal of Molecular Sciences 18, no. 7: 1350. https://doi.org/10.3390/ijms18071350





