Profiling the Extended Cleavage Specificity of the House Dust Mite Protease Allergens Der p 1, Der p 3 and Der p 6 for the Prediction of New Cell Surface Protein Substrates
Abstract
:1. Introduction
2. Results
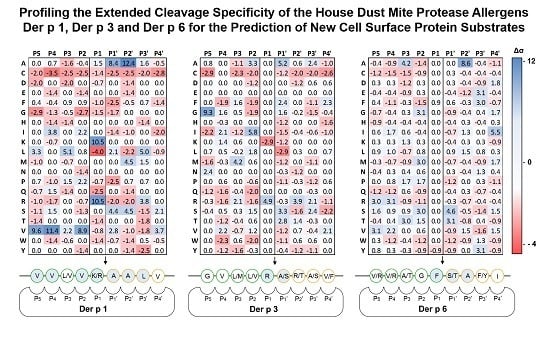
2.1. Determination of Extended Der p 1, Der p 3 and Der p 6 Substrate Specificities
2.2. In Silico Prediction of Potential Protein Substrates within the Human Cell Surface Proteome
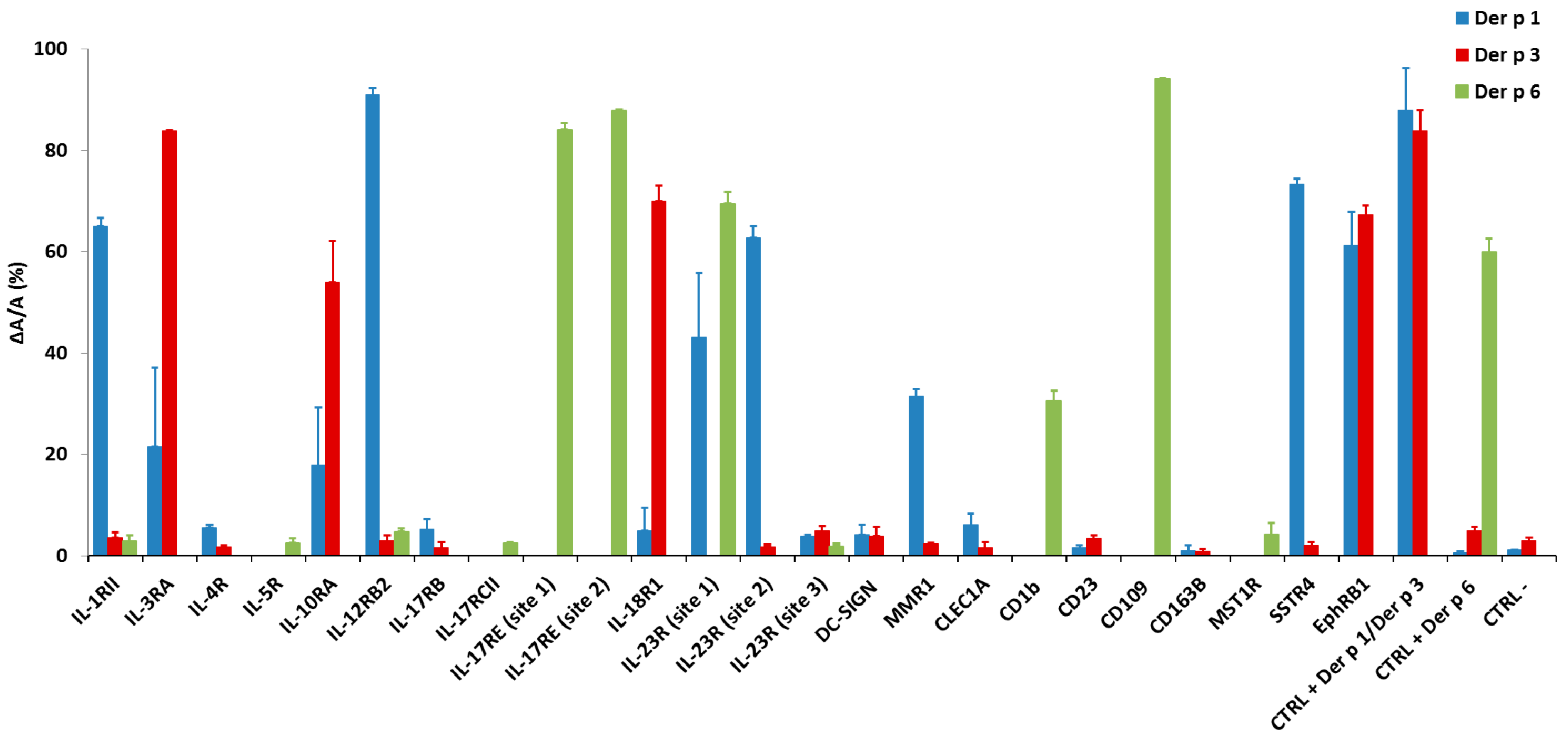
2.3. Validation of the Predictions by ELISA Using Phage Peptide Substrates
2.4. Validation of the Interleukin-23 Receptor as a Specific Target for Der p 1 and Der p 6
3. Discussion
4. Materials and Methods
4.1. Expression, Purification and Maturation of Recombinant Der p 1, Der p 3 and Der p 6
4.2 Construction of Phage Substrate Libraries
4.3. Screening of Phage-Substrate Libraries
4.4. Determination of Der p 1, Der p 3 and Der p 6 Substrate Specificity Profiles and Prediction of New Protein Substrates among the Human Cell Surface Proteome
4.5. Cleavage Site Validation by Phage ELISA
4.6. In Vitro Cleavage of the Recombinant Interleukin-23 Receptor by HDM Proteases
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CD1b | T-cell surface glycoprotein CD1b |
| CD23 | Low-affinity immunoglobulin epsilon Fc receptor |
| CD109 | 150 kDa TGF-β-1-binding protein CD109 antigen |
| CD163b | Scavenger receptor cysteine-rich type 1 protein M160 |
| CLEC1A | C-type lectin domain family 1 member A |
| DC-SIGN | Dendritic cell-specific |
| EphRB1 | Ephrin type B receptor 1 |
| HRP | Horseradish peroxidase. ICAM-3-grabbing non-integrin 1 |
| IL | Interleukin |
| R | Receptor |
| A | α |
| B | β |
| II | Type 2 |
| MMR1 | Macrophage mannose receptor 1 |
| MST1R | Macrophage-stimulating protein receptor |
| SSTR4 | Somatostatin receptor type 4 |
References
- Jacquet, A. Innate immune responses in house dust mite allergy. ISRN Allergy 2013, 2013, 735031. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.R. Hierarchy and molecular properties of house dust mite allergens. Allergol. Int. 2015, 64, 304–311. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.D.; Rowntree, S.; Mitchell, E.B.; de Fuenmajor, M.C.P.; Platts-Mills, T.A. Quantitative assessments of IgG and IgE antibodies to inhalant allergens in patients with atopic dermatitis. J. Allergy Clin. Immunol. 1983, 72, 27–33. [Google Scholar] [CrossRef]
- Platts-Mills, T.A.; Chapman, M.D. Dust mites: Immunology, allergic disease, and environmental control. J. Allergy Clin. Immunol. 1987, 80, 755–775. [Google Scholar] [CrossRef]
- Herman, J.; Thelen, N.; Smargiasso, N.; Mailleux, A.C.; Luxen, A.; Cloes, M.; de Pauw, E.; Chevigne, A.; Galleni, M.; Dumez, M.E. Der p 1 is the primary activator of Der p 3, Der p 6 and Der p 9 the proteolytic allergens produced by the house dust mite Dermatophagoides pteronyssinus. Biochim. Biophys. Acta 2014, 1840, 1117–1124. [Google Scholar] [CrossRef] [PubMed]
- Dumez, M.E.; Herman, J.; Campizi, V.; Galleni, M.; Jacquet, A.; Chevigne, A. Orchestration of an uncommon maturation cascade of the house dust mite protease allergen quartet. Front. Immunol. 2014, 5, 138. [Google Scholar] [CrossRef] [PubMed]
- Dumez, M.E.; Teller, N.; Mercier, F.; Tanaka, T.; Vandenberghe, I.; Vandenbranden, M.; Devreese, B.; Luxen, A.; Frere, J.M.; Matagne, A.; et al. Activation mechanism of recombinant Der p 3 allergen zymogen: Contribution of cysteine protease Der p 1 and effect of propeptide glycosylation. J. Biol. Chem. 2008, 283, 30606–30617. [Google Scholar] [CrossRef] [PubMed]
- Bouaziz, A.; Walgraffe, D.; Bouillot, C.; Herman, J.; Foguenne, J.; Gothot, A.; Louis, R.; Hentges, F.; Jacquet, A.; Mailleux, A.C.; et al. Development of recombinant stable house dust mite allergen Der p 3 molecules for component-resolved diagnosis and specific immunotherapy. Clin. Exp. Allergy 2015, 45, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Hales, B.J.; Martin, A.C.; Pearce, L.J.; Laing, I.A.; Hayden, C.M.; Goldblatt, J.; Le Souef, P.N.; Thomas, W.R. IgE and IgG anti-house dust mite specificities in allergic disease. J. Allergy Clin. Immunol. 2006, 118, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Yasueda, H.; Mita, H.; Akiyama, K.; Shida, T.; Ando, T.; Sugiyama, S.; Yamakawa, H. Allergens from dermatophagoides mites with chymotryptic activity. Clin. Exp. Allergy 1993, 23, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, P.T.; Elliot, C.E.; Lenzo, J.C.; Jarnicki, A.G.; Larcombe, A.N.; Zosky, G.R.; Holt, P.G.; Thomas, W.R. Sensitizing and TH2 adjuvant activity of cysteine protease allergens. Int. Arch. Allergy Immunol. 2012, 158, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Deb, R.; Shakib, F.; Reid, K.; Clark, H. Major house dust mite allergens Dermatophagoides pteronyssinus 1 and Dermatophagoides farinae 1 degrade and inactivate lung surfactant proteins A and D. J. Biol. Chem. 2007, 282, 36808–36819. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Winton, H.L.; Soeller, C.; Tovey, E.R.; Gruenert, D.C.; Thompson, P.J.; Stewart, G.A.; Taylor, G.W.; Garrod, D.R.; Cannell, M.B.; et al. Der p 1 facilitates transepithelial allergen delivery by disruption of tight junctions. J. Clin. Investig. 1999, 104, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Iijima, K.; Elias, M.K.; Seno, S.; Tojima, I.; Kobayashi, T.; Kephart, G.M.; Kurabayashi, M.; Kita, H. Airway uric acid is a sensor of inhaled protease allergens and initiates type 2 immune responses in respiratory mucosa. J. Immunol. 2014, 192, 4032–4042. [Google Scholar] [CrossRef] [PubMed]
- Kool, M.; Willart, M.A.; van Nimwegen, M.; Bergen, I.; Pouliot, P.; Virchow, J.C.; Rogers, N.; Osorio, F.; Reis e Sousa, C.; Hammad, H.; et al. An unexpected role for uric acid as an inducer of T helper 2 cell immunity to inhaled antigens and inflammatory mediator of allergic asthma. Immunity 2011, 34, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Takai, T.; Fujimura, T.; Matsuoka, H.; Ogawa, T.; Murayama, K.; Ishii, A.; Ikeda, S.; Okumura, K.; Ogawa, H. Mite serine protease activates protease-activated receptor-2 and induces cytokine release in human keratinocytes. Allergy 2009, 64, 1366–1374. [Google Scholar] [CrossRef] [PubMed]
- Adam, E.; Hansen, K.K.; Astudillo Fernandez, O.; Coulon, L.; Bex, F.; Duhant, X.; Jaumotte, E.; Hollenberg, M.D.; Jacquet, A. The house dust mite allergen Der p 1, unlike Der p 3, stimulates the expression of interleukin-8 in human airway epithelial cells via a proteinase-activated receptor-2-independent mechanism. J. Biol. Chem. 2006, 281, 6910–6923. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.; Stacey, M.A.; Schmidt, M.; Mori, L.; Mattoli, S. Interaction of mite allergens Der p 3 and Der p 9 with protease-activated receptor-2 expressed by lung epithelial cells. J. Immunol. 2001, 167, 1014–1021. [Google Scholar] [CrossRef] [PubMed]
- Furmonaviciene, R.; Ghaemmaghami, A.M.; Boyd, S.E.; Jones, N.S.; Bailey, K.; Willis, A.C.; Sewell, H.F.; Mitchell, D.A.; Shakib, F. The protease allergen Der p 1 cleaves cell surface DC-SIGN and DC-SIGNR: Experimental analysis of in silico substrate identification and implications in allergic responses. Clin. Exp. Allergy 2007, 37, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.; Sutton, B.J.; Beavil, R.L.; Shi, J.; Sewell, H.F.; Gould, H.J.; Laing, P.; Shakib, F. Cleavage of the low-affinity receptor for human IgE (CD23) by a mite cysteine protease: Nature of the cleaved fragment in relation to the structure and function of CD23. Eur. J. Immunol 1997, 27, 584–588. [Google Scholar] [CrossRef] [PubMed]
- Ghaemmaghami, A.M.; Shakib, F. Human t cells that have been conditioned by the proteolytic activity of the major dust mite allergen Der p 1 trigger enhanced immunoglobulin E synthesis by B cells. Clin. Exp. Allergy 2002, 32, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.; Sewell, H.F.; Shakib, F. Proteolytic cleavage of CD25, the α subunit of the human T cell interleukin 2 receptor, by Der p 1, a major mite allergen with cysteine protease activity. J. Exp. Med. 1998, 187, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Kalsheker, N.A.; Deam, S.; Chambers, L.; Sreedharan, S.; Brocklehurst, K.; Lomas, D.A. The house dust mite allergen Der p 1 catalytically inactivates α 1-antitrypsin by specific reactive centre loop cleavage: A mechanism that promotes airway inflammation and asthma. Biochem. Biophys. Res. Commun. 1996, 221, 59–61. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Farmer, K.; MacDonald, L.; Kalsheker, N.; Pritchard, D.; Haslett, C.; Lamb, J.; Sallenave, J.M. House dust mite Der p 1 downregulates defenses of the lung by inactivating elastase inhibitors. Am. J. Respir. Cell Mol. Biol. 2003, 29, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Herbert, C.A.; King, C.M.; Ring, P.C.; Holgate, S.T.; Stewart, G.A.; Thompson, P.J.; Robinson, C. Augmentation of permeability in the bronchial epithelium by the house dust mite allergen Der p 1. Am. J. Respir. Cell Mol. Biol. 1995, 12, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Takai, T.; Kuhara, T.; Ota, M.; Kato, T.; Hatanaka, H.; Ichikawa, S.; Tokura, T.; Akiba, H.; Mitsuishi, K.; et al. Crucial commitment of proteolytic activity of a purified recombinant major house dust mite allergen Der p 1 to sensitization toward IgE and IgG responses. J. Immunol. 2006, 177, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.; Laing, P.; Sewell, H.F.; Shakib, F. Der p I, a major allergen of the house dust mite, proteolytically cleaves the low-affinity receptor for human IgE (CD23). Eur. J. Immunol. 1995, 25, 3191–3194. [Google Scholar] [CrossRef] [PubMed]
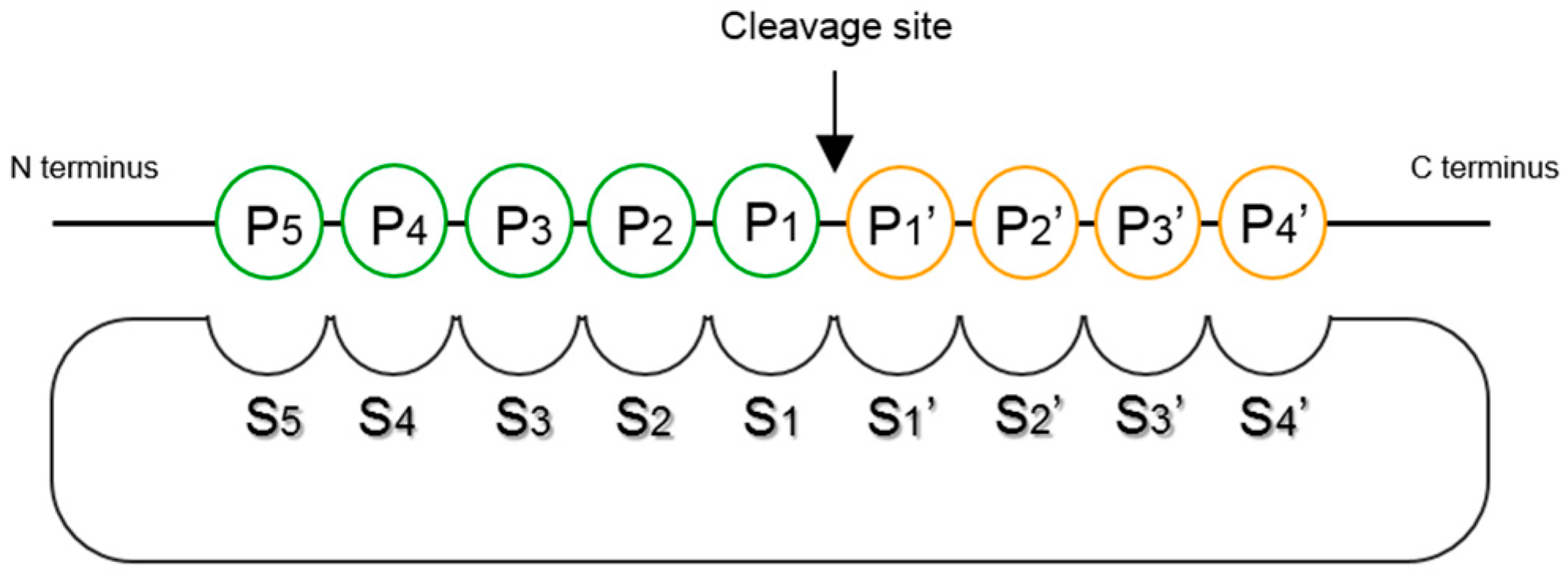
- Schechter, I.; Berger, A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967, 27, 157–162. [Google Scholar] [CrossRef]
- Harris, J.; Mason, D.E.; Li, J.; Burdick, K.W.; Backes, B.J.; Chen, T.; Shipway, A.; van Heeke, G.; Gough, L.; Ghaemmaghami, A.; et al. Activity profile of dust mite allergen extract using substrate libraries and functional proteomic microarrays. Chem. Biol. 2004, 11, 1361–1372. [Google Scholar] [CrossRef] [PubMed]
- Schulz, O.; Sewell, H.F.; Shakib, F. A sensitive fluorescent assay for measuring the cysteine protease activity of Der p 1, a major allergen from the dust mite Dermatophagoides pteronyssinus. Mol. Pathol. 1998, 51, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Dall'Acqua, W.; Halin, C.; Rodrigues, M.L.; Carter, P. Elastase substrate specificity tailored through substrate-assisted catalysis and phage display. Protein Eng. 1999, 12, 981–987. [Google Scholar] [CrossRef] [PubMed]
- Iida, H.; Takai, T.; Hirasawa, Y.; Kamijo, S.; Shimura, S.; Ochi, H.; Nishioka, I.; Maruyama, N.; Ogawa, H.; Okumura, K.; et al. Epicutaneous administration of papain induces IgE and IgG responses in a cysteine protease activity-dependent manner. Allergol. Int. 2014, 63, 219–226. [Google Scholar] [CrossRef] [PubMed]
- De Boer, J.D.; Van’t Veer, C.; Stroo, I.; van der Meer, A.J.; de Vos, A.F.; van der Zee, J.S.; Roelofs, J.J.; van der Poll, T. Protease-activated receptor-2 deficient mice have reduced house dust mite-evoked allergic lung inflammation. Innate Immun. 2014, 20, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Post, S.; Heijink, I.H.; Petersen, A.H.; de Bruin, H.G.; van Oosterhout, A.J.; Nawijn, M.C. Protease-activated receptor-2 activation contributes to house dust mite-induced IgE responses in mice. PLoS ONE 2014, 9, e91206. [Google Scholar] [CrossRef] [PubMed]
- Newton, G.K.; Perrior, T.R.; Jenkins, K.; Major, M.R.; Key, R.E.; Stewart, M.R.; Firth-Clark, S.; Lloyd, S.M.; Zhang, J.; Francis-Newton, N.J.; et al. The discovery of potent, selective, and reversible inhibitors of the house dust mite peptidase allergen Der p 1: An innovative approach to the treatment of allergic asthma. J. Med. Chem. 2014, 57, 9447–9462. [Google Scholar] [CrossRef] [PubMed]
- Matthews, D.J.; Goodman, L.J.; Gorman, C.M.; Wells, J.A. A survey of furin substrate specificity using substrate phage display. Protein Sci. 1994, 3, 1197–1205. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.M.; Shi, L.; Navre, M. Rapid identification of highly active and selective substrates for stromelysin and matrilysin using bacteriophage peptide display libraries. J. Biol. Chem. 1995, 270, 6440–6449. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, S.; Miyadera, K.; Sugimoto, Y.; Matsuo, K.; Wierzba, K.; Yamada, Y. Substrate phage as a tool to identify novel substrate sequences of proteases. Comb. Chem. High Throughput Screen. 2001, 4, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Delhalle, S.; Schmit, J.C.; Chevigne, A. Phages and HIV-1: From display to interplay. Int. J. Mol. Sci. 2012, 13, 4727–4794. [Google Scholar] [CrossRef] [PubMed]
- Choe, Y.; Leonetti, F.; Greenbaum, D.C.; Lecaille, F.; Bogyo, M.; Bromme, D.; Ellman, J.A.; Craik, C.S. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006, 281, 12824–12832. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.J.; Thomas, W.R. Cloning and sequencing of the group 6 allergen of Dermatophagoides pteronyssinus. Clin. Exp. Allergy 1996, 26, 1150–1154. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.; Potempa, J.; Travis, J. Inactivation of α 1-proteinase inhibitor as a broad screen for detecting proteolytic activities in unknown samples. Anal. Biochem. 1998, 260, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Chevigne, A.; Barumandzadeh, R.; Groslambert, S.; Cloes, B.; Dehareng, D.; Filee, P.; Marx, J.C.; Frere, J.M.; Matagne, A.; Jacquet, A.; et al. Relationship between propeptide pH unfolding and inhibitory ability during ProDer p 1 activation mechanism. J. Mol. Biol. 2007, 374, 170–185. [Google Scholar] [CrossRef] [PubMed]
- Gallwitz, M.; Enoksson, M.; Thorpe, M.; Hellman, L. The extended cleavage specificity of human thrombin. PLoS ONE 2012, 7, e31756. [Google Scholar] [CrossRef] [PubMed]
- Bock, P.E.; Panizzi, P.; Verhamme, I.M. Exosites in the substrate specificity of blood coagulation reactions. J. Thromb. Haemost. 2007, 5 (Suppl. S1), 81–94. [Google Scholar] [CrossRef] [PubMed]
- Chevigne, A.; Campizi, V.; Szpakowska, M.; Bourry, D.; Dumez, M.E.; Martins, J.C.; Matagne, A.; Galleni, M.; Jacquet, A. The Lys-Asp-Tyr triad within the mite allergen Der p 1 propeptide is a critical structural element for the pH-dependent initiation of the protease maturation. Int. J. Mol. Sci. 2017, 18, 1087. [Google Scholar] [CrossRef] [PubMed]
- Szabo, A.; Salameh, M.A.; Ludwig, M.; Radisky, E.S.; Sahin-Toth, M. Tyrosine sulfation of human trypsin steers S2′ subsite selectivity towards basic amino acids. PLoS ONE 2014, 9, e102063. [Google Scholar] [CrossRef] [PubMed]
- Beckham, S.A.; Boyd, S.E.; Reynolds, S.; Willis, C.; Johnstone, M.; Mika, A.; Simerska, P.; Wijeyewickrema, L.C.; Smith, A.I.; Kemp, D.J.; et al. Characterization of a serine protease homologous to house dust mite group 3 allergens from the scabies mite sarcoptes scabiei. J. Biol. Chem. 2009, 284, 34413–34422. [Google Scholar] [CrossRef] [PubMed]
- Hamza, T.; Barnett, J.B.; Li, B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010, 11, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Palomares, O.; Martin-Fontecha, M.; Lauener, R.; Traidl-Hoffmann, C.; Cavkaytar, O.; Akdis, M.; Akdis, C.A. Regulatory T cells and immune regulation of allergic diseases: Roles of IL-10 and TGF-β. Genes Immun. 2014, 15, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Nishikado, H.; Fujimura, T.; Taka, H.; Mineki, R.; Ogawa, H.; Okumura, K.; Takai, T. Cysteine protease antigens cleave CD123, the α subunit of murine IL-3 receptor, on basophils and suppress IL-3-mediated basophil expansion. Biochem. Biophys. Res. Commun. 2015, 460, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Tworek, D.; Smith, S.G.; Salter, B.M.; Baatjes, A.J.; Scime, T.; Watson, R.; Obminski, C.; Gauvreau, G.M.; O’Byrne, P.M. IL-25 receptor expression on airway dendritic cells after allergen challenge in subjects with asthma. Am. J. Respir. Crit. Care Med. 2016, 193, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.; Iraola, V.; Leonor, J.R.; Carnes, J. Enzymatic activity of allergenic house dust and storage mite extracts. J. Med. Entomol. 2013, 50, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Parmley, S.F.; Smith, G.P. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene 1988, 73, 305–318. [Google Scholar] [CrossRef]
- Cwirla, S.E.; Peters, E.A.; Barrett, R.W.; Dower, W.J. Peptides on phage: A vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. USA 1990, 87, 6378–6382. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.L.; Peterson, E.P.; Hudig, D.; Thornberry, N.A.; Craik, C.S. Definition and redesign of the extended substrate specificity of granzyme b. J. Biol. Chem. 1998, 273, 27364–27373. [Google Scholar] [CrossRef] [PubMed]
- Boyd, S.E.; Pike, R.N.; Rudy, G.B.; Whisstock, J.C.; de la Banda, M.G. Pops: A computational tool for modeling and predicting protease specificity. J. Bioinform. Comput. Biol. 2005, 3, 551–585. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.Y.; Park, H.R.; Shin, J.H.; Kim, S.W.; Cho, J.H.; Park, Y.J.; Kim, S.W. The serine protease inhibitor, 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride, reduces allergic inflammation in a house dust mite allergic rhinitis mouse model. Allergy Asthma Immunol. Res. 2014, 6, 558–566. [Google Scholar] [CrossRef] [PubMed]


| Protease | Library | Displayed Sequence | P1 Residue |
|---|---|---|---|
| Der p 1 | X4-X-X4 | His6-GPGG-X4-X-X4-GGPG-pIII | X |
| Der p 3 | X4-R/K-X4 | His6-GPGG-X4-R/K-X4-GGPG-pIII | R/K |
| Der p 6 | X4-Y/F-X4 | His6-GPGG-X4-Y/F-X4-GGPG-pIII | Y/F |
| Target | Proteases | Predicted Cleavage Site P5P4P3P2P1↓P1′P2′P3′P4′ | Cleavage Position | Extracellular Domain | Uniprot |
|---|---|---|---|---|---|
| DC-SIGN | Der p 1 | LVVIK↓SAEE | 295−296 | 59−404 | Q9NNX6 |
| IL-1RII | Der p 1 | PVALR↓CPQV | 49−50 | 14−343 | P27930 |
| IL-12RBII | Der p 1 | AVAVS↓AANS | 398−399 | 24−622 | Q99665 |
| IL-17RB | Der p 1 | KKCVK↓AGSL | 175−176 | 18−292 | Q9NRM6 |
| IL-23R (site 2) | Der p 1 | VVHVK↓SLET | 163−164 | 24−355 | Q5VWK5 |
| CLEC1A | Der p 1 | VQNIK↓LAGS | 109−110 | 74−280 | Q8NC01 |
| CD163b | Der p 1 | RVEVK↓HADT | 811−812 | 41−1359 | Q9NR16 |
| IL-3RA | Der p 1, Der p 3 | LVRGR↓SAAF | 187−188 | 19−305 | P26951 |
| IL-4R | Der p 1, Der p 3 | HVKPR↓APGN | 124−125 | 26−232 | P24394 |
| IL-10RA | Der p 1, Der p 3 | GYRAR↓VRAV | 99−100 | 22−235 | Q13651 |
| IL-18R1 | Der p 1, Der p 3 | ILVRK↓ADMA | 315−316 | 22−319 | Q13478 |
| EphRB1 | Der p 1, Der p 3 | VVQVR↓ARTV | 507−508 | 18−540 | P54762 |
| IL-23R (site 1) | Der p 1, Der p 6 | LVWVQ↓AANA | 197−198 | 24−355 | Q5VWK5 |
| CD23 | Der p 3 | QLEER↓AARN | 59−60 | 48−321 | P06734 |
| IL-23R (site 3) | Der p 3 | AVISR↓AETI | 227−228 | 24−355 | Q5VWK5 |
| MMR1 | Der p 3 | PGGRR↓SSLS | 1042−1043 | 19−1389 | P22897 |
| SSTR4 | Der p 3 | PGDAR↓AAGM | 43−44 | 1−46 | P31391 |
| IL-5RαII | Der p 6 | LHKGF↓SASV | 94−95 | 21−342 | Q01344 |
| IL-17RCII | Der p 6 | VVLSF↓QAYP | 200−201 | 21−538 | Q8NAC3 |
| IL-17RE (site 1) | Der p 6 | SFTGS↓SAYI | 47−55 | 24−454 | Q8NRF9 |
| IL-17RE (site 2) | Der p 6 | MHATF↓SAAW | 386−387 | 24−454 | Q8NRF9 |
| CD1b | Der p 6 | RAQKF↓CALI | 162−163 | 18−303 | P29016 |
| CD109 | Der p 6 | EDGSF↓SAFG | 974−975 | 22−1420 | Q6YHK3 |
| MST1R | Der p 6 | VVPSF↓SAGG | 49−50 | 25−297 | Q04912 |
| (A) | |||
| Target | Cleavage Site for Der p 1 | ΔA/A (%) | Shared |
| IL-12RBII | AVAVS↓AANS | 90.9 ± 1.4 | – |
| SSTR4 | PGDAR↓AAGM | 73.4 ± 1.1 | – |
| IL-1RII | PVALR↓CPQV | 65.0 ± 1.8 | Der p 6 |
| IL-23R (site 2) | VVHVK↓SLET | 62.8 ± 12.7 | – |
| EphRB1 | VVQVR↓ARTV | 61.1 ± 6.7 | Der p 3 |
| IL-23R (site 1) | LVWVQ↓AANA | 43.1 ± 2.2 | Der p 6 |
| MMR1 | PGGRR ↓SSLS | 31.6 ± 1.3 | – |
| IL-3RA | LVRGR↓SAAF | 21.5 ± 15.5 | Der p 3 |
| IL-10RA | GYRAR↓VRAV | 17.9 ± 11.3 | Der p 3 |
| CLEC1A | VQNIK↓LAGS | 6.1 ± 2.12 | – |
| IL-4R | HVKPR↓APGN | 5.5 ± 0.5 | – |
| IL-17RB | KKCVK↓AGSL | 5.2 ± 2.5 | – |
| IL-18R1 | FILVRK↓ADMA | 4.9 ± 4.52 | Der p 3 |
| DC-SIGN | LVVIK↓SAEE | 4.2 ± 1.9 | – |
| IL-23R (site 3) | AVISR↓AETI | 3.8 ± 0.3 | Der p 6 |
| Ctrl neg | GGSGGSGGS | 1.15 ± 0.1 | – |
| (B) | |||
| Target | Cleavage Site for Der p 3 | ΔA/A (%) | Shared |
| IL-3RA | LVRGR↓SAAF | 83.8 ± 0.2 | Der p 1 |
| IL-18R1 | FILVRK↓ADMA | 70.0 ± 9 | Der p 1 |
| EphRB1 | VVQVR↓ARTV | 67.2 ± 1.7 | Der p 1 |
| IL-10RA | GYRAR↓VRAV | 54.0 ± 8.0 | Der p 1 |
| Ctrl neg | GGSGGSGGS | 3.0 ± 0.6 | – |
| (C) | |||
| Target | Cleavage Site for Der p 6 | ΔA/A (%) | Shared |
| CD109 | EDGSF↓SAFG | 94.1 ± 0.1 | – |
| IL-17RE (site 2) | MHATF↓SAAW | 87.9 ± 0.1 | – |
| IL-17RE (site 1) | SFTGS↓SAYI | 84.1 ± 1.2 | – |
| IL-23R (site 1) | LVWVQ↓AANA | 69.4 ± 2.3 | Der p 1 |
| CD1b | RAQKF↓CALI | 30.4 ± 2.0 | – |
| IL-12RBII | AVAVS↓AANS | 4.7 ± 0.6 | – |
| MST1R | VVPSF↓SAGG | 4.3 ± 2.1 | – |
| IL-1RII | PVALR↓CPQV | 2.9 ± 1.1 | Der p 1 |
| IL-5RαII | LHKGF↓SASV | 2.5 ± 0.8 | – |
| IL-17RCII | VVLSF↓QAYP | 2.4 ± 0.2 | – |
| IL-23R (site 3) | AVISR↓AETI | 1.9 ± 0.4 | Der p 1 |
| Ctrl neg | GGSGGSGGS | 0.0 ± 0.0 | – |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacquet, A.; Campisi, V.; Szpakowska, M.; Dumez, M.-E.; Galleni, M.; Chevigné, A. Profiling the Extended Cleavage Specificity of the House Dust Mite Protease Allergens Der p 1, Der p 3 and Der p 6 for the Prediction of New Cell Surface Protein Substrates. Int. J. Mol. Sci. 2017, 18, 1373. https://doi.org/10.3390/ijms18071373
Jacquet A, Campisi V, Szpakowska M, Dumez M-E, Galleni M, Chevigné A. Profiling the Extended Cleavage Specificity of the House Dust Mite Protease Allergens Der p 1, Der p 3 and Der p 6 for the Prediction of New Cell Surface Protein Substrates. International Journal of Molecular Sciences. 2017; 18(7):1373. https://doi.org/10.3390/ijms18071373
Chicago/Turabian StyleJacquet, Alain, Vincenzo Campisi, Martyna Szpakowska, Marie-Eve Dumez, Moreno Galleni, and Andy Chevigné. 2017. "Profiling the Extended Cleavage Specificity of the House Dust Mite Protease Allergens Der p 1, Der p 3 and Der p 6 for the Prediction of New Cell Surface Protein Substrates" International Journal of Molecular Sciences 18, no. 7: 1373. https://doi.org/10.3390/ijms18071373