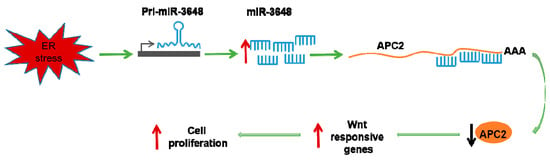
Induction of miR-3648 Upon ER Stress and Its Regulatory Role in Cell Proliferation
Abstract
:1. Introduction
2. Results
2.1. Expression of miR-3648 in Human Cell Lines and Tissues
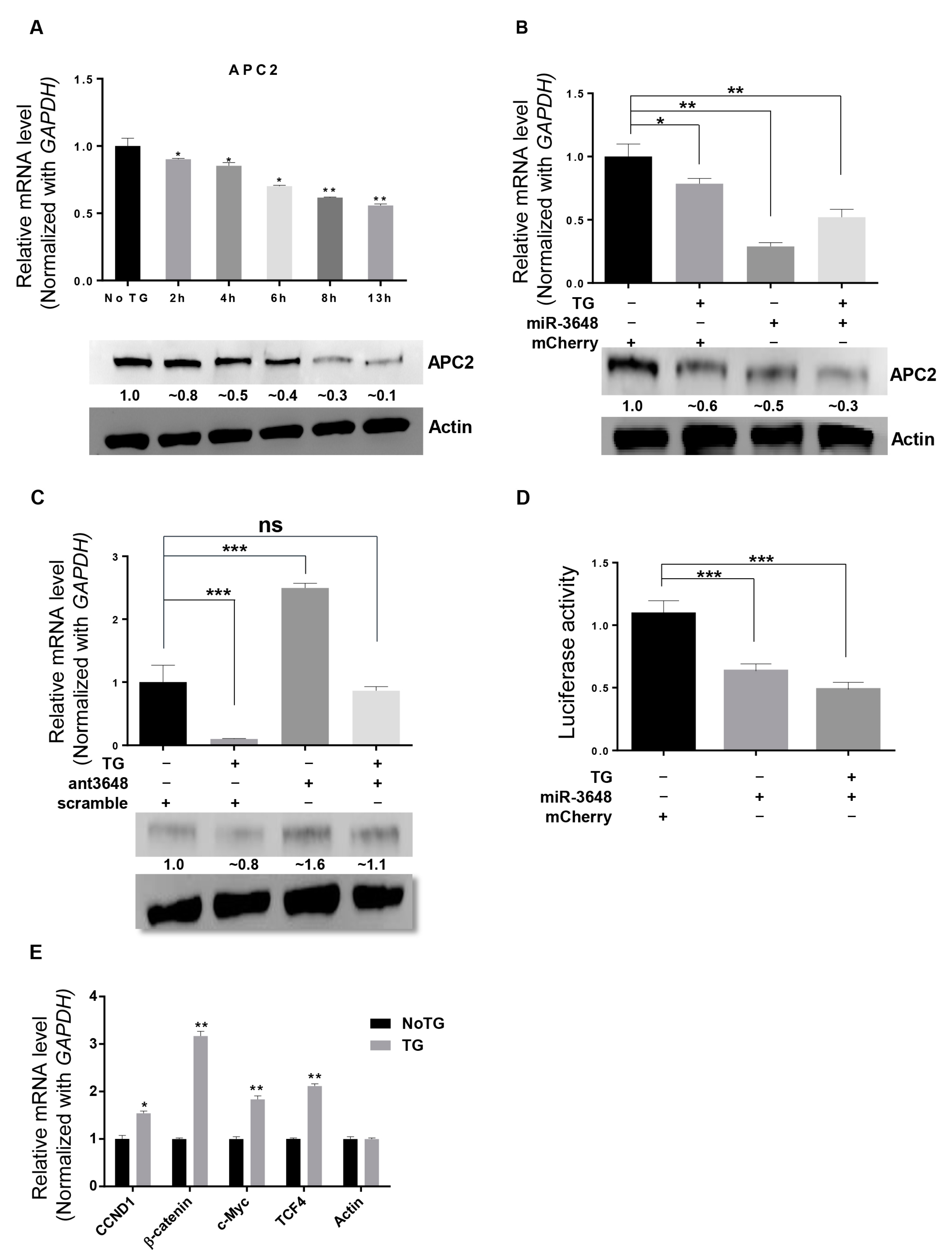
2.2. miR-3648 Was Induced by the ER Stress
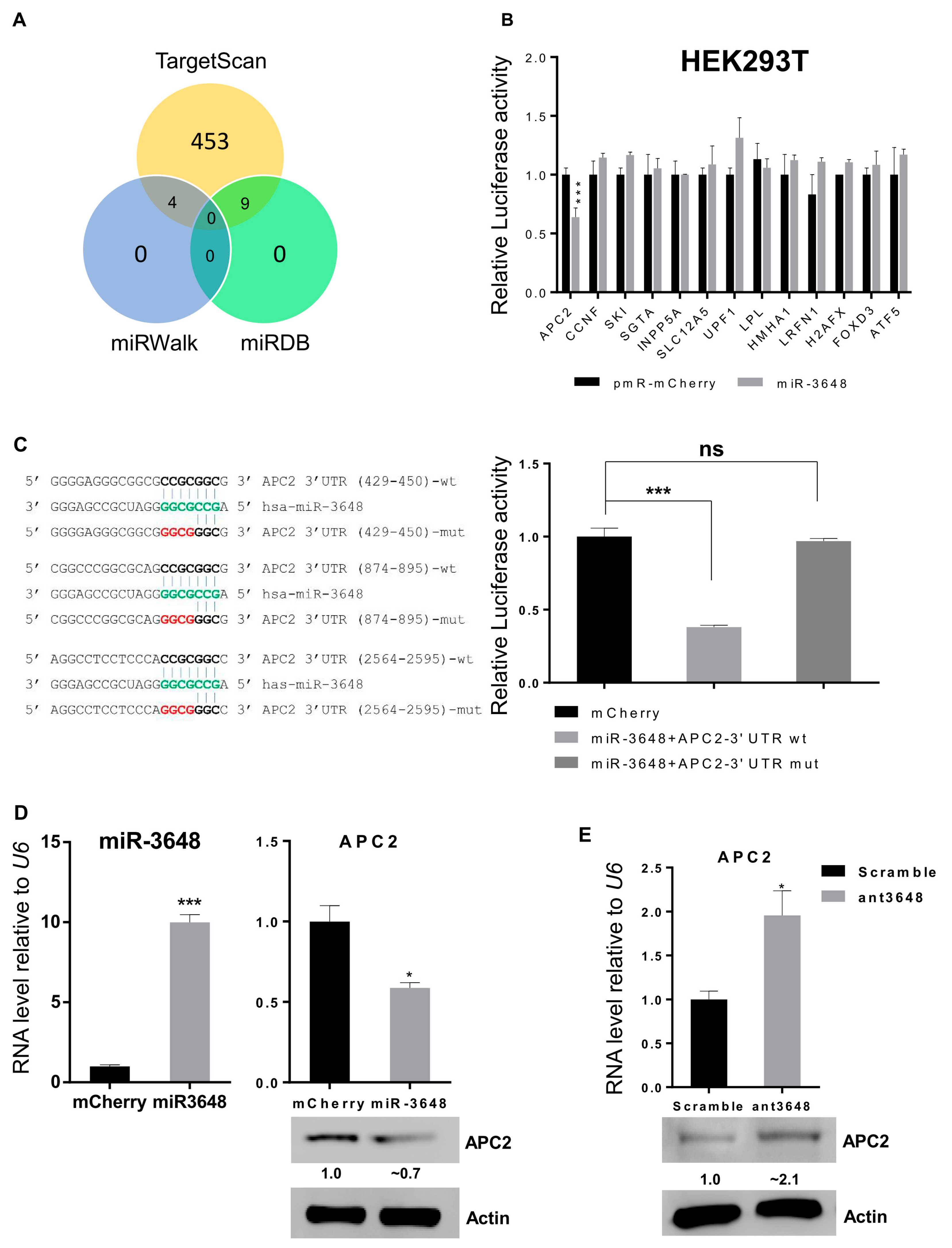
2.3. miR-3648 Directly Targeted the 3′ UTR of APC2
2.4. APC2 Was Regulated by miR-3648 under ER Stress
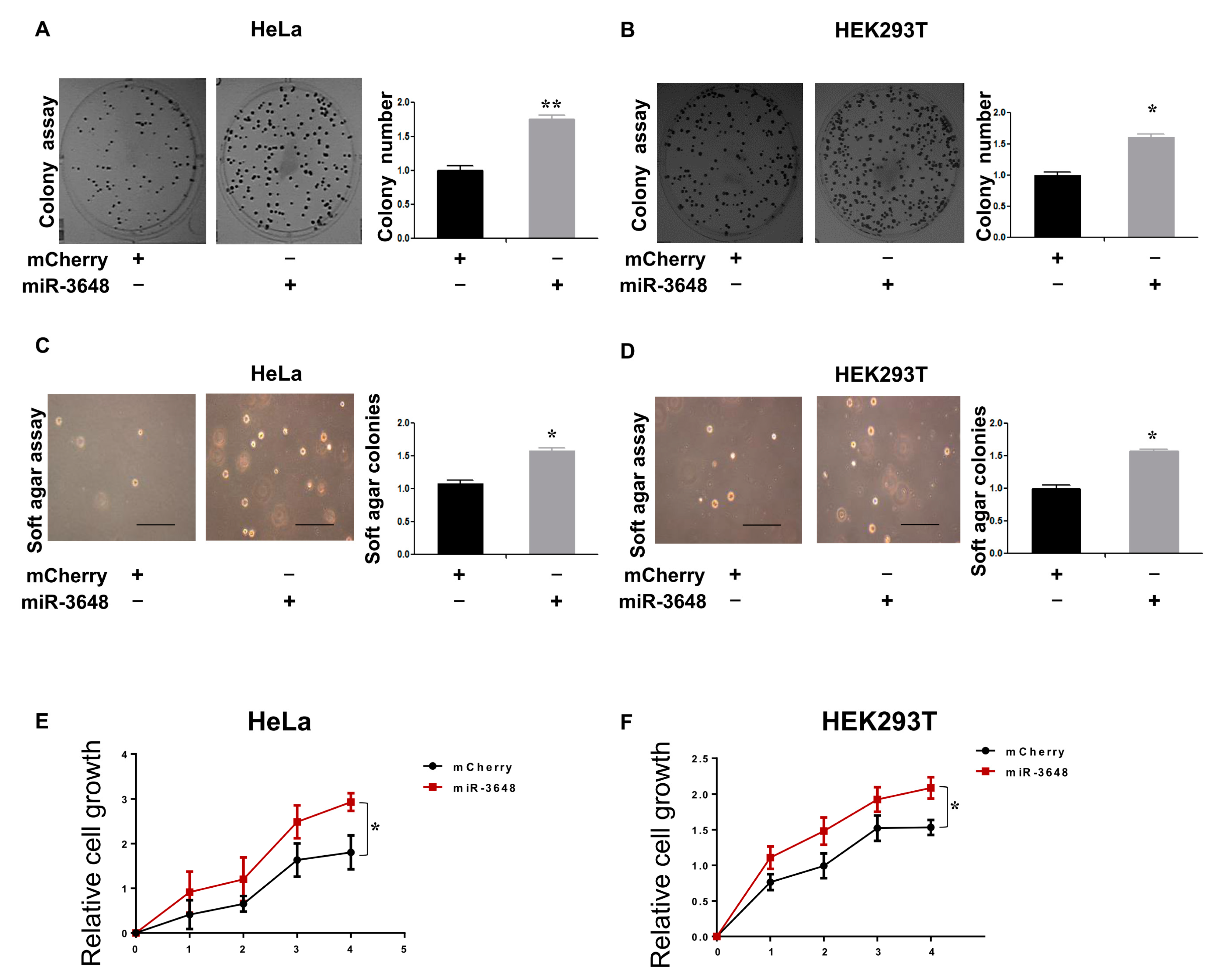
2.5. Upregulation of miR-3648 Increased Cell Proliferation
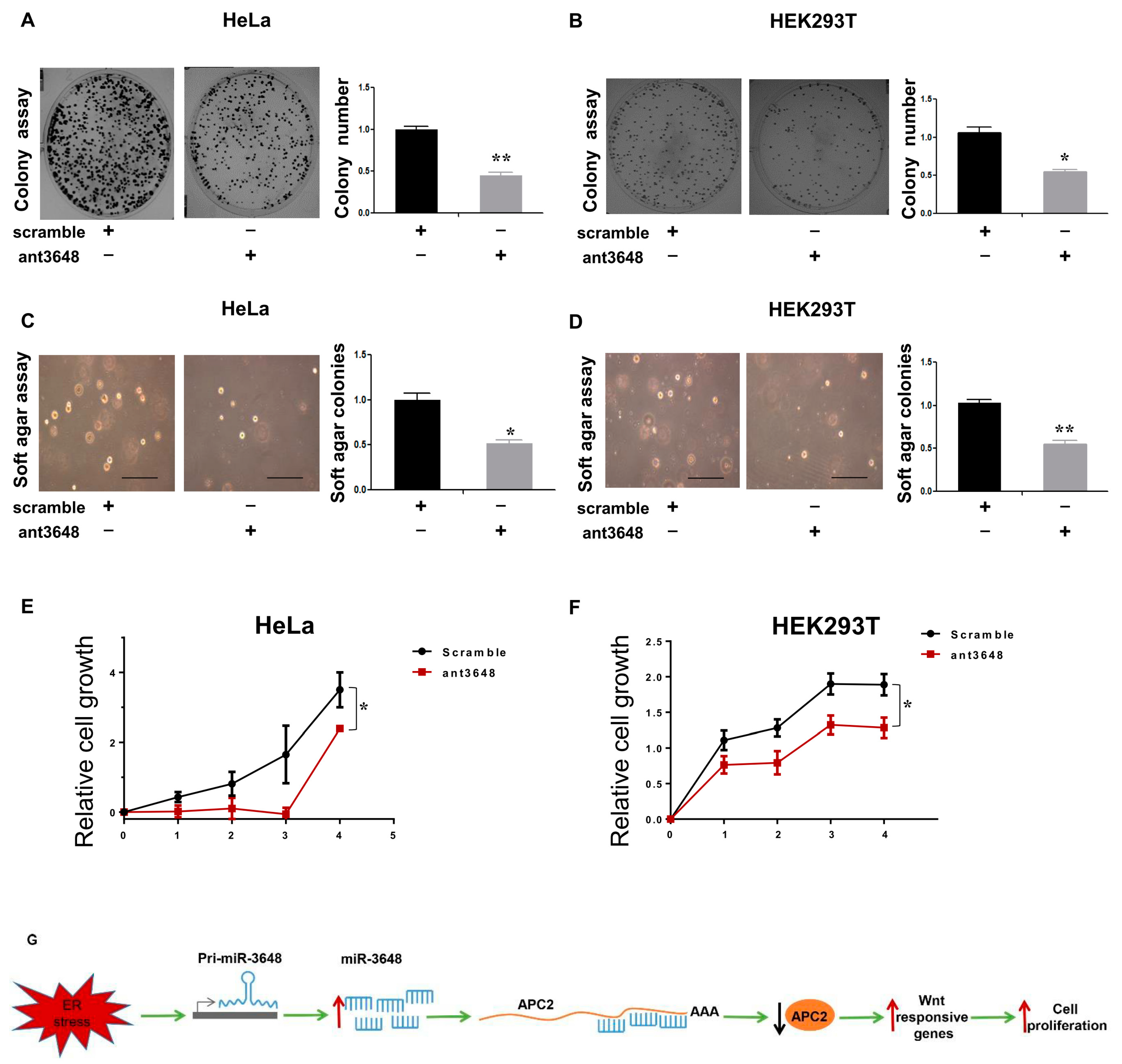
2.6. Suppression of miR-3648 Decreased Cell Proliferation
3. Discussion
4. Materials and Methods
4.1. Construction of Plasmids
4.2. Cell Culture, Transfections, TG and TM Treatments
4.3. PCR Reactions
4.4. miRNA Isolation and Northern Blot
4.5. Luciferase Assay
4.6. Western Blots
4.7. Colonogenic Assay
4.8. Soft Agar Assay
4.9. Cell Proliferation Assay
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| APC2 | Adenomatous polyposis coli 2 |
| ER | Endoplasmic Reticulum |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| TG | Thapsigargin |
| TM | Tunicamycin |
References
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Pehserl, A.M.; Ress, A.L.; Stanzer, S.; Resel, M.; Karbiener, M.; Stadelmeyer, E.; Stiegelbauer, V.; Gerger, A.; Mayr, C.; Scheideler, M.; et al. Comprehensive Analysis of miRNome Alterations in Response to Sorafenib Treatment in Colorectal Cancer Cells. Int. J. Mol. Sci. 2016, 17, 2011. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, functionand decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene regulation by non-coding RNAs. Crit Rev. Biochem. Mol. Biol. 2013, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Schetter, A.J.; Leung, S.Y.; Sohn, J.J.; Zanetti, K.A.; Bowman, E.D.; Yanaihara, N.; Yuen, S.T.; Chan, T.L.; Kwong, D.L.; Au, G.K.; et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA 2008, 299, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Chitnis, N.S.; Pytel, D.; Bobrovnikova-Marjon, E.; Pant, D.; Zheng, H.; Maas, N.L.; Frederick, B.; Kushner, J.A.; Chodosh, L.A.; Koumenis, C.; et al. MiR-211 is a prosurvival microRNA that regulates chop expression in a PERK dependent manner. Mol. Cell 2012, 48, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Muratsu-Ikeda, S.; Nangaku, M.; Ikeda, Y.; Tanaka, T.; Wada, T.; Inagi, R. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS ONE 2012, 7, e41462. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.V.; Kupai, K.; Szucs, G.; Gaspar, R.; Paloczi, J.; Farago, N.; Zvara, A.; Puskás, L.G.; Rázga, Z.; Tiszlavicz, L.; et al. MicroRNA-25-dependent up-regulation of NADPH oxidase4 (NOX4) mediates hypercholesterolemia-induced oxidative/nitrative stress and subsequent dysfunction in the heart. J. Mol. Cell. Cardiol. 2013, 62, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Diehl, J.A.; Fuchs, S.Y.; Koumenis, C. The cell biology of the unfolded protein response. Gastroenterology 2011, 141, 38–41. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Lawrence, D.A.; Marsters, S.; Acosta-Alvear, D.; Kimmig, P.; Mendez, A.S.; Paton, A.W.; Paton, J.C.; Walter, P.; Ashkenazi, A. Opposing unfolded-protein-response signals converge on death receptor 5 to control apoptosis. Science 2014, 345, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Dorner, A.J.; Wasley, L.C.; Kaufman, R.J. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992, 11, 1563–1571. [Google Scholar] [PubMed]
- Melnick, J.; Aviel, S.; Argon, Y. The endoplasmic reticulum stress protein GRP94, in addition to BiP, associates with unassembled immunoglobulin chains. J. Biol. Chem. 1992, 267, 21303–21306. [Google Scholar] [PubMed]
- Li, W.; Hsiung, W.; Zhou, Y.; Roy, B.; Lee, A.S. Induction of the mammalian GRP78/BiP gene by Ca2+ depletion and formation of aberrant proteins: Activation of the conserved stress-inducible grp core promoter element by the human nuclear factor YY1. Mol. Cell. Biol. 1997, 17, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Wang, L.; Wang, B.; Wang, T.; Yang, G.; Shen, L.; Wang, T.; Guo, X.; Liu, Y.; Xia, Y.; et al. Activation of volume-sensitive outwardly rectifying chloride channel by ROS contributes to ER stress and cardiac contractile dysfunction: Involvement of CHOP through Wnt. Cell Death Dis. 2014, 5, e1528. [Google Scholar] [CrossRef]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Hetz, C.; Martinon, F.; Rodriguez, D.; Glimcher, L.H. The unfolded protein response: Integrating stress signals through the stress sensor IRE1 α. Physiol. Rev. 2011, 91, 1219–1243. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Haze, K.; Yanagi, H.; Yura, T.; Mori, K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998, 273, 33741–33749. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Baumeister, P.; Roy, B.; Phan, T.; Foti, D.; Luo, S.; Lee, A.S. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 2000, 20, 5096–5106. [Google Scholar] [CrossRef] [PubMed]
- Schroder, M.; Kaufman, R.J. The mammalian unfolded protein response. Annu. Rev. Biochem. 2005, 74, 739–789. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Reed, J.C. ER stress-induced cell death mechanisms. Biochim. Biophysi. Acta-Mol. Cell Res. 2013, 1833, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller-Herzog, C.; Hall, M.N. Bidirectional crosstalk between endoplasmic reticulum stress and mTOR signaling. Trends Cell Boil. 2012, 22, 274–282. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, L.; Pei, C.; Lin, X.; Yuan, Z. Dysfunction of Wntless triggers the retrograde Golgi-to-ER transport of Wingless and induces ER stress. Sci. Rep. 2015, 6, 19418. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Huang, C.; Chen, L.; Shan, G.; Li, Z. Altered expression of microRNAs in the response to ER stress. Sci. Bull. 2015, 60, 202–209. [Google Scholar] [CrossRef]
- Dokanehiifard, S.; Yasari, A.; Najafi, H.; Jafarzadeh, M.; Nikkhah, M.; Mowla, S.J.; Soltani, B.M. A novel microRNA located in TrkC gene regulates Wnt signaling pathway and is differentially expressed in colorectal cancer specimens. J. Biol. Chem. 2017, M116, 760710. [Google Scholar] [CrossRef] [PubMed]
- Schneikert, J.; Harsha, S.; Chandra, V.; Ruppert, J.G.; Ray, S.; Wenzel, E.M.; Behrens, J. Functional Comparison of Human Adenomatous Polyposis Coli (APC) and APC-Like in Targeting β-Catenin for Degradation. PLoS ONE 2013, 8, e68072. [Google Scholar] [CrossRef] [PubMed]
- Daly, C.S.; Shaw, P.; Ordonez, L.D.; Williams, G.T.; Quist, J.; Grigoriadis, A.; Van Es, J.H.; Clevers, H.; Clarke, A.R.; Reed, K.R. Functional redundancy between Apc and Apc2 regulates tissue homeostasis and prevents tumorigenesis in murine mammary epithelium. Oncogene 2016, 36, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, H.; Murata, Y.; Koyama, K.; Fujiyama, A.; Miyoshi, Y.; Monden, M.; Akiyama, T.; Nakamura, Y. Identification of a Brain-specific APC Homologue, APCL, and Its Interaction with/β-Catenin. Cancer. Res. 1998, 58, 5176–5181. [Google Scholar] [PubMed]
- Ying, X.; Li-ya, Q.; Feng, Z.; Yin, W.; Ji-hong, L. MiR-939 promotes the proliferation of human ovarian cancer cells by repressing APC2 expression. Biomed. Pharmacother. 2015, 71, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.Y.; He, L.; Fominykh, K.; Yan, Z.; Guo, S.; Zhang, X.; Taylor, M.S.; Tang, L.; Li, J.; Liu, J.; et al. Evolution of the human-specific microRNA miR-941. Nat. Commun. 2012, 3, 1145. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Bu, Y.; Chitnis, N.; Koumenis, C.; Fuchs, S.Y.; Diehl, J.A. miR-216b regulation of c-Jun mediates GADD153/CHOP-dependent apoptosis. Nat. Commun. 2016, 7, 11422. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, K.; Hall, C.; Sato, K.; Lairmore, T.; Marzioni, M.; Glaser, S.; Meng, F.; Alpini, G. Lin28 and let-7: Roles and regulation in liver diseases. Am. J. Physiol. Gastrointest. Liver Physiol. 2016, 310, G757–G765. [Google Scholar] [CrossRef] [PubMed]
- Shan, G.; Li, Y.; Zhang, J.; Li, W.; Szulwach, K.E.; Duan, R.; Faghihi, M.A.; Khalil, A.M.; Lu, L.; Paroo, Z.; et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008, 26, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. miRWalk-database: Prediction of possible miRNA binding sites by “walking” the genes of 3 genomes. J. Biomed. Inform. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Nathan, W.; Xiaowei, W. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015, 43, 146–152. [Google Scholar] [CrossRef]
- Zaravinos, A.; Lambrou, G.I.; Mourmouras, N.; Katafygiotis, P.; Papagregoriou, G.; Giannikou, K.; Delakas, D.; Deltas, C. New miRNA Profiles Accurately Distinguish Renal Cell Carcinomas and Upper Tract Urothelial Carcinomas from the Normal Kidney. PLoS ONE 2014, 9, e91646. [Google Scholar] [CrossRef] [PubMed]
- Kumari, B.; Jain, P.; Das, S.; Ghosal, S.; Hazra, B.; Trivedi, A.C.; Basu, A.; Chakrabarti, J.; Vrati, S.; Banerjee, A. Dynamic changes in global microRNAome and transcriptome reveal complex miRNA-mRNA regulated host response to Japanese Encephalitis Virus in microglial cells. Sci. Rep. 2016, 6, 20263. [Google Scholar] [CrossRef] [PubMed]
- Morishita, A.; Iwama, H.; Fujihara, S.; Sakamoto, T.; Fujita, K.; Tani, J.; Miyoshi, H.; Yoneyama, H.; Himoto, T.; Masaki, T. MicroRNA profiles in various hepatocellular carcinoma cell lines. Oncol. Lett. 2016, 12, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Tardif, K.D.; Waris, G.; Siddiqui, A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005, 13, 159–163. [Google Scholar] [CrossRef]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell. Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef]
- Ochoa-Hernandez, A.B.; Juarez-Vazquez, C.I.; Rosales-Reynoso, M.A.; Barros-Nunez, P. Wnt-β-catenin signaling pathway and its relationship with cancer. Cir. Cir. 2012, 80, 389–398. [Google Scholar] [PubMed]
- Kim, K.H.; Seol, H.J.; Kim, E.H.; Rheey, J.; Jin, H.J.; Lee, Y.; Joo, K.M.; Lee, J.; Nam, D.H. Wnt/β-catenin signaling is a key downstream mediator of met signaling in glioblastoma stem cells. Neuro. Oncol. 2013, 15, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, I.S.; Chang, K.C.; Tsai, Y.T.; Ke, J.Y.; Lu, P.J.; Lee, K.H.; Yeh, S.D.; Hong, T.M.; Chen, Y.L. Microrna-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the wnt/β-catenin signaling pathway. Carcinogenesis 2013, 34, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Awan, H.M.; Shah, A.; Rashid, F.; Shan, G. Primate specific Long Non-coding RNAs and MicroRNAs. Genom. Proteom. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, F.; Awan, H.M.; Shah, A.; Chen, L.; Shan, G. Induction of miR-3648 Upon ER Stress and Its Regulatory Role in Cell Proliferation. Int. J. Mol. Sci. 2017, 18, 1375. https://doi.org/10.3390/ijms18071375
Rashid F, Awan HM, Shah A, Chen L, Shan G. Induction of miR-3648 Upon ER Stress and Its Regulatory Role in Cell Proliferation. International Journal of Molecular Sciences. 2017; 18(7):1375. https://doi.org/10.3390/ijms18071375
Chicago/Turabian StyleRashid, Farooq, Hassaan Mehboob Awan, Abdullah Shah, Liang Chen, and Ge Shan. 2017. "Induction of miR-3648 Upon ER Stress and Its Regulatory Role in Cell Proliferation" International Journal of Molecular Sciences 18, no. 7: 1375. https://doi.org/10.3390/ijms18071375