Transgenic Strategies for Sparse but Strong Expression of Genetically Encoded Voltage and Calcium Indicators
Abstract
:1. Introduction
2. Results
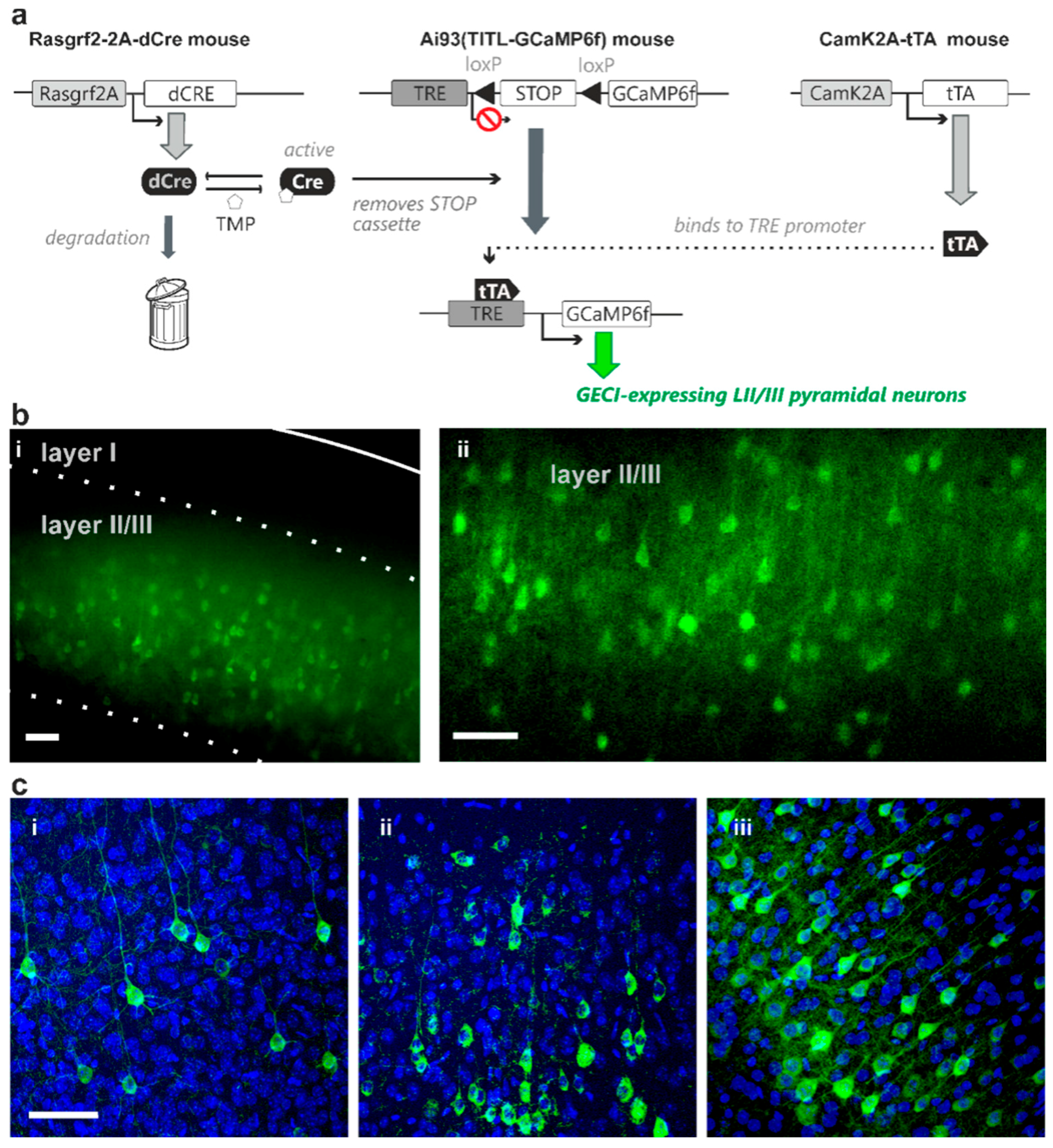
2.1. TMP Dose-Dependent Control of dCre-Mediated Recombination
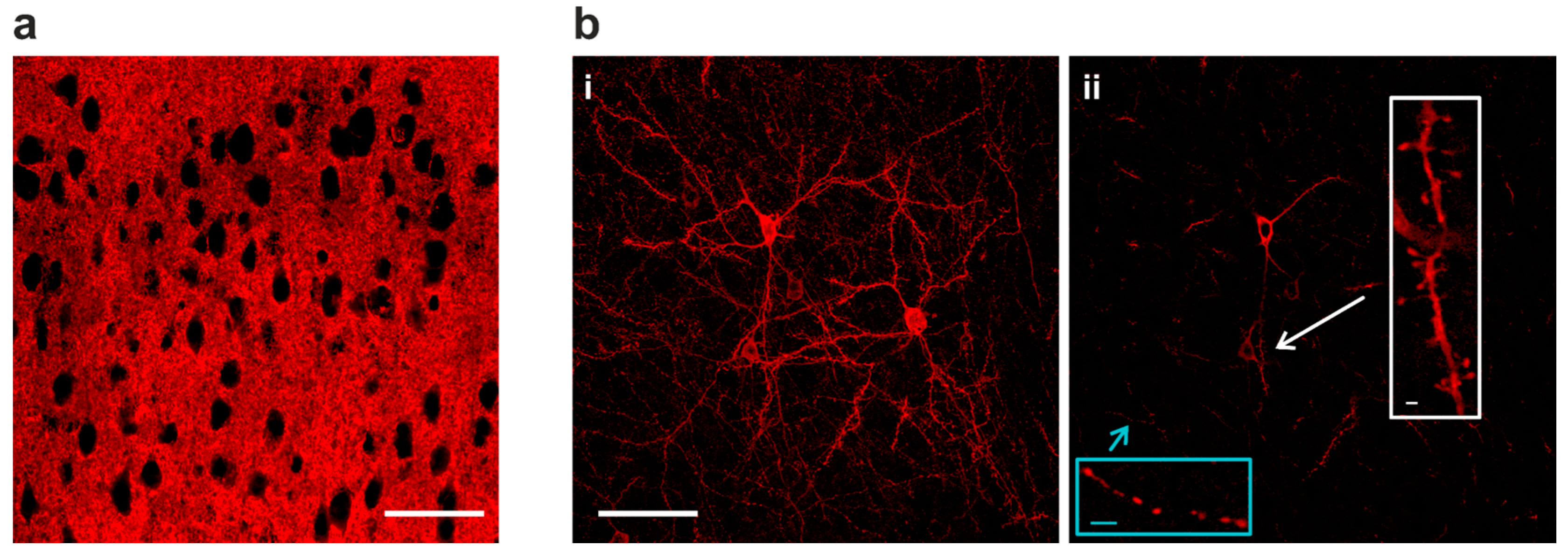
2.2. Sparse Expression of GEVI Uncovers the Morphologies of Individual Cells
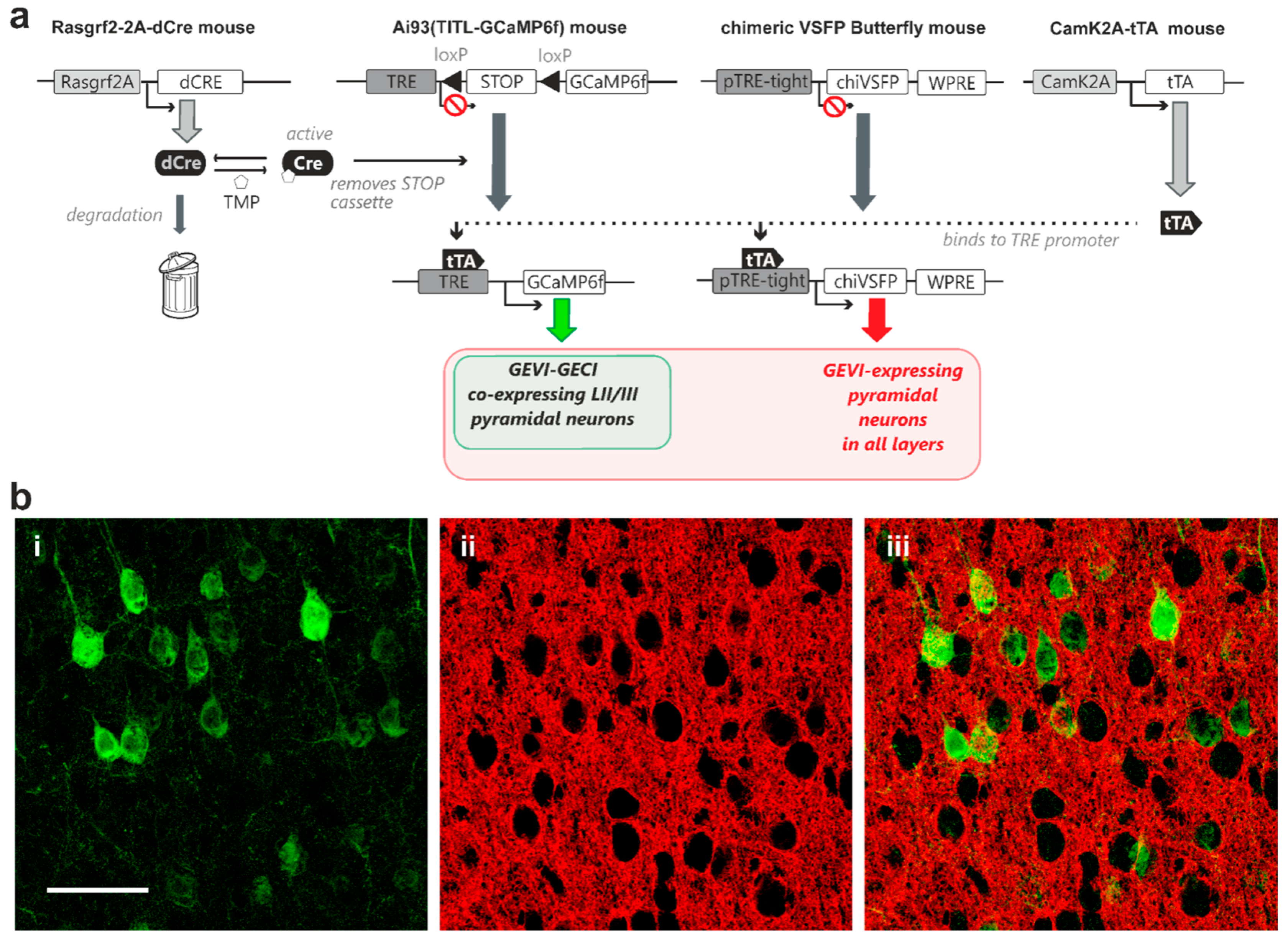
2.3. Modular Transgenic Strategies Allow Controlled Co-Expression of GECI and GEVI
3. Discussion
3.1. GEVI and GECI Transgenic Approach
3.2. Sparse Strong GEVI Expression Resolves Individual Cells and Subcellular Structures
3.3. GEVI/GECI Co-Expression
4. Materials and Methods
4.1. Animals
4.2. Inducible Control of Recombination Using Trimethoprim
4.3. Histology and Confocal Imaging
4.4. Ex Vivo Quantification of Indicator Expression
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Knöpfel, T. Genetically encoded optical indicators for the analysis of neuronal circuits. Nat. Rev. Neurosci. 2012, 13, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Antic, S.D.; Empson, R.M.; Knöpfel, T. Voltage imaging to understand connections and functions of neuronal circuits. J. Neurophysiol 2016, 116, 135–152. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.Z.; Schnitzer, M.J. Genetically encoded indicators of neuronal activity. Nat. Neurosci. 2016, 19, 1142–1153. [Google Scholar] [CrossRef] [PubMed]
- Knöpfel, T.; Diez-Garcia, J.; Akemann, W. Optical probing of neuronal circuit dynamics: Genetically encoded versus classical fluorescent sensors. Trends Neurosci. 2006, 29, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Barnes, S.; Knöpfel, T. Mammalian cortical voltage imaging using genetically encoded voltage indicators: A review honoring professor amiram grinvald. Neurophotonics 2017, 4, 031214. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.A.; Hirokawa, K.E.; Sorensen, S.A.; Gu, H.; Mills, M.; Ng, L.L.; Bohn, P.; Mortrud, M.; Ouellette, B.; Kidney, J.; et al. Anatomical characterization of cre driver mice for neural circuit mapping and manipulation. Front. Neural Circuits 2014, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.J.; Zeng, H. Genetic approaches to neural circuits in the mouse. Annu. Rev. Neurosci. 2013, 36, 183–215. [Google Scholar] [CrossRef] [PubMed]
- Madisen, L.; Garner, A.R.; Shimaoka, D.; Chuong, A.S.; Klapoetke, N.C.; Li, L.; van der Bourg, A.; Niino, Y.; Egolf, L.; Monetti, C.; et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 2015, 85, 942–958. [Google Scholar] [CrossRef] [PubMed]
- Sando, R., III; Baumgaertel, K.; Pieraut, S.; Torabi-Rander, N.; Wandless, T.J.; Mayford, M.; Maximov, A. Inducible control of gene expression with destabilized Cre. Nat. Methods 2013, 10, 1085–1088. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Akemann, W.; Mutoh, H.; Perron, A.; Park, Y.K.; Iwamoto, Y.; Knöpfel, T. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J. Neurophysiol 2012, 108, 2323–2337. [Google Scholar] [CrossRef] [PubMed]
- Mishina, Y.; Mutoh, H.; Song, C.; Knöpfel, T. Exploration of genetically encoded voltage indicators based on a chimeric voltage sensing domain. Front. Mol. Neurosci. 2014, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Shen, E.H.; Hohmann, J.G.; Oh, S.W.; Bernard, A.; Royall, J.J.; Glattfelder, K.J.; Sunkin, S.M.; Morris, J.A.; Guillozet-Bongaarts, A.L.; et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 2012, 149, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Mayford, M.; Bach, M.E.; Huang, Y.Y.; Wang, L.; Hawkins, R.D.; Kandel, E.R. Control of memory formation through regulated expression of a camkii transgene. Science 1996, 274, 1678–1683. [Google Scholar] [CrossRef] [PubMed]
- Empson, R.M.; Goulton, C.; Scholtz, D.; Gallero-Salas, Y.; Zeng, H.; Knöpfel, T. Validation of optical voltage reporting by the genetically encoded voltage indicator VSFP-butterfly from cortical layer 2/3 pyramidal neurons in mouse brain slices. Physiol. Rep. 2015, 3, e12468. [Google Scholar] [CrossRef] [PubMed]
- Akemann, W.; Sasaki, M.; Mutoh, H.; Imamura, T.; Honkura, N.; Knöpfel, T. Two-photon voltage imaging using a genetically encoded voltage indicator. Sci. Rep. 2013, 3, 2231. [Google Scholar] [CrossRef] [PubMed]
- Akemann, W.; Mutoh, H.; Perron, A.; Rossier, J.; Knöpfel, T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat. Methods 2010, 7, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.D.; Li, J.Z.; Zhang, Y.; Gong, Y.; St-Pierre, F.; Lin, M.Z.; Schnitzer, M.J. Cell-type-specific optical recording of membrane voltage dynamics in freely moving mice. Cell 2016, 167, 1650–1662. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, F.; Marshall, J.D.; Yang, Y.; Gong, Y.; Schnitzer, M.J.; Lin, M.Z. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci. 2014, 17, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Huang, C.; Li, J.Z.; Grewe, B.F.; Zhang, Y.; Eismann, S.; Schnitzer, M.J. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 2015, 350, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Wekselblatt, J.B.; Flister, E.D.; Piscopo, D.M.; Niell, C.M. Large-scale imaging of cortical dynamics during sensory perception and behavior. J. Neurophysiol. 2016, 115, 2852–2866. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Horie, K.; Madisen, L.; Pavlova, M.N.; Gragerova, G.; Rohde, A.D.; Schimpf, B.A.; Liang, Y.; Ojala, E.; Kramer, F.; et al. An inducible and reversible mouse genetic rescue system. PLoS Genet. 2008, 4, e1000069. [Google Scholar] [CrossRef] [PubMed]
- Carandini, M.; Shimaoka, D.; Rossi, L.F.; Sato, T.K.; Benucci, A.; Knöpfel, T. Imaging the awake visual cortex with a genetically encoded voltage indicator. J. Neurosci. 2015, 35, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Shimaoka, D.; Song, C.; Knöpfel, T. State-dependent modulation of slow wave motifs towards awakening. Front. Cell Neurosci. 2017, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Madisen, L.; Zwingman, T.A.; Sunkin, S.M.; Oh, S.W.; Zariwala, H.A.; Gu, H.; Ng, L.L.; Palmiter, R.D.; Hawrylycz, M.J.; Jones, A.R.; et al. A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010, 13, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Madisen, L.; Mao, T.; Koch, H.; Zhuo, J.M.; Berenyi, A.; Fujisawa, S.; Hsu, Y.W.; Garcia, A.J., III; Gu, X.; Zanella, S.; et al. A toolbox of cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat. Neurosci. 2012, 15, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Knöpfel, T.; Gallero-Salas, Y.; Song, C. Genetically encoded voltage indicators for large scale cortical imaging come of age. Curr. Opin. Chem. Biol. 2015, 27, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Wzorek, J.S.; Knöpfel, T.F.; Sapountzis, I.; Evans, D.A. A macrocyclic approach to tetracycline natural products. Investigation of transannular alkylations and michael additions. Org. Lett. 2012, 14, 5840–5843. [Google Scholar] [CrossRef] [PubMed]
- Grinvald, A.; Hildesheim, R. Vsdi: A new era in functional imaging of cortical dynamics. Nat. Rev. Neurosci. 2004, 5, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Wagner, M.J.; Zhong Li, J.; Schnitzer, M.J. Imaging neural spiking in brain tissue using fret-opsin protein voltage sensors. Nat. Commun. 2014, 5, 3674. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Zhao, Y.; Douglass, A.D.; Hochbaum, D.R.; Brinks, D.; Werley, C.A.; Harrison, D.J.; Campbell, R.E.; Cohen, A.E. Bright and fast multicoloured voltage reporters via electrochromic fret. Nat. Commun. 2014, 5, 4625. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.; Monuki, E.S.; Tang, H.; Imitola, J.; Haubst, N.; Khoury, S.J.; Cunningham, J.; Gotz, M.; Walsh, C.A. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. J. Comp. Neurol. 2004, 479, 168–180. [Google Scholar] [CrossRef] [PubMed]
- Milojkovic, B.A.; Zhou, W.L.; Antic, S.D. Voltage and calcium transients in basal dendrites of the rat prefrontal cortex. J. Physiol. 2007, 585, 447–468. [Google Scholar] [CrossRef] [PubMed]
- Canepari, M.; Djurisic, M.; Zecevic, D. Dendritic signals from rat hippocampal CA1 pyramidal neurons during coincident pre- and post-synaptic activity: A combined voltage- and calcium-imaging study. J. Physiol. 2007, 580, 463–484. [Google Scholar] [CrossRef] [PubMed]

| (TMP) (mg/kg Body Weight) | No. of Animals | Total No. of Sections | Expressing Cells/All Nuclei | Scaled Expression Probability |
|---|---|---|---|---|
| 750 | 5 | 25 | 25.29 ± 1.88% | 1.00 |
| 0.03 | 5 | 25 | 20.02 ± 0.83% | 0.79 |
| 0.005 | 2 | 10 | 15.68 ± 2.63% | 0.62 |
| 0.001 | 2 | 4 | 4.51 ± 1.06% | 0.18 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, C.; Do, Q.B.; Antic, S.D.; Knöpfel, T. Transgenic Strategies for Sparse but Strong Expression of Genetically Encoded Voltage and Calcium Indicators. Int. J. Mol. Sci. 2017, 18, 1461. https://doi.org/10.3390/ijms18071461
Song C, Do QB, Antic SD, Knöpfel T. Transgenic Strategies for Sparse but Strong Expression of Genetically Encoded Voltage and Calcium Indicators. International Journal of Molecular Sciences. 2017; 18(7):1461. https://doi.org/10.3390/ijms18071461
Chicago/Turabian StyleSong, Chenchen, Quyen B. Do, Srdjan D. Antic, and Thomas Knöpfel. 2017. "Transgenic Strategies for Sparse but Strong Expression of Genetically Encoded Voltage and Calcium Indicators" International Journal of Molecular Sciences 18, no. 7: 1461. https://doi.org/10.3390/ijms18071461