Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection
Abstract
:1. Introduction
2. Results
2.1. The Sequence Analysis of ChiIV3 and Its Promoter pChiIV3
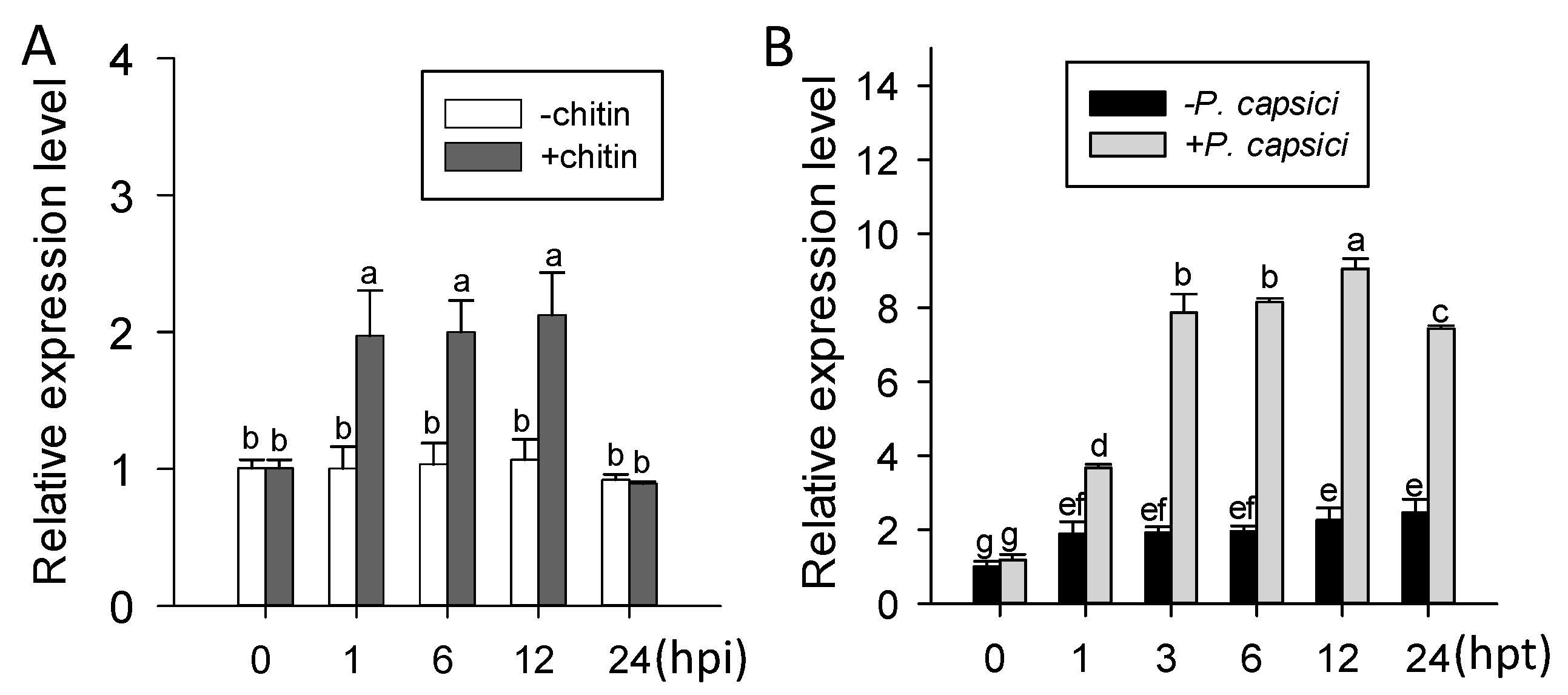
2.2. ChiIV3 Was Transcriptionally Induced in Pepper Plants against Phytophthora capsici Inoculation (PCI) and Applied Chitin Treatment
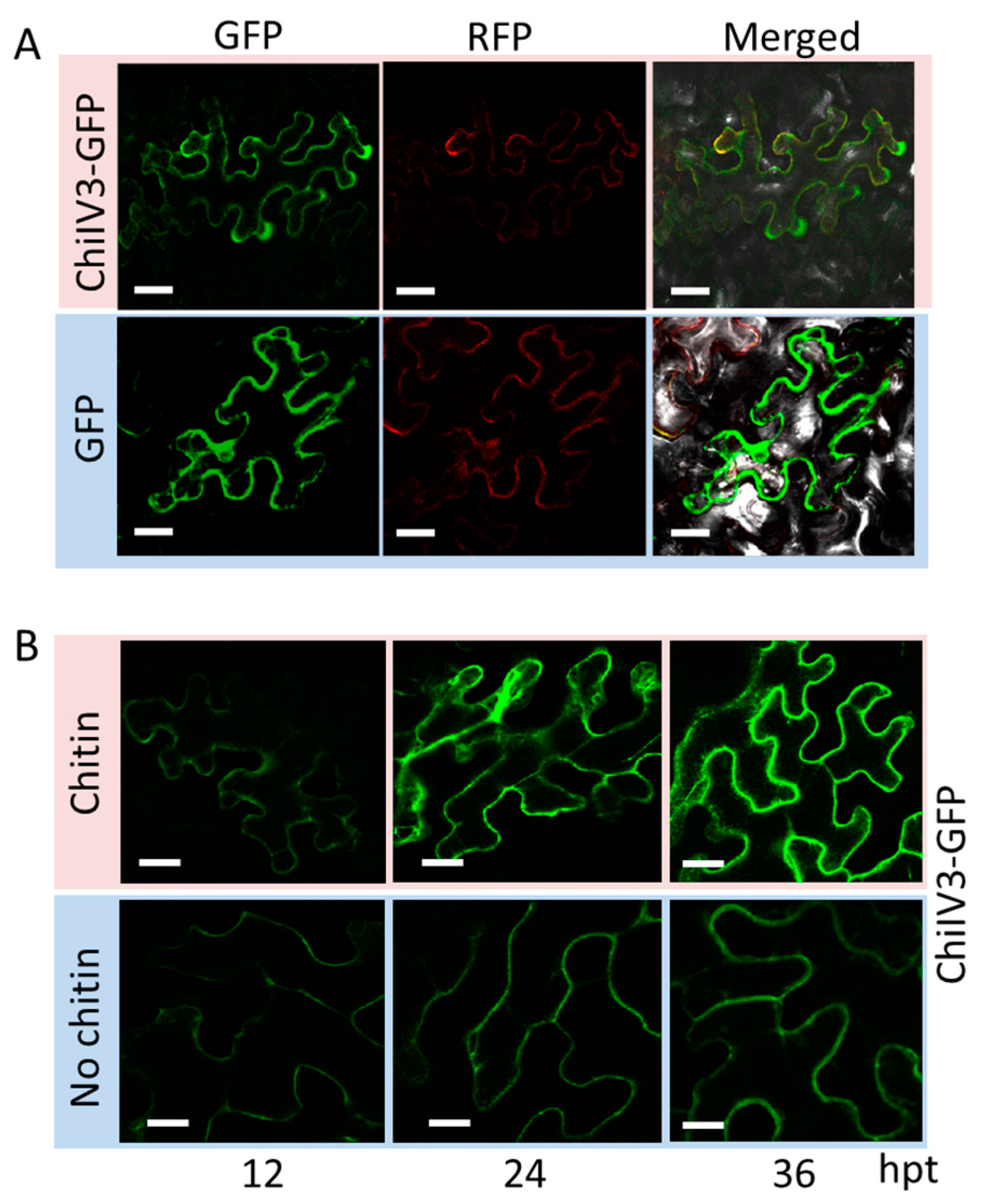
2.3. ChiIV3 Protein Is Localized to the Plasma Membrane and Can Be Activated by Applied Chitin Treatment in Nicotiana benthamiana Leaves
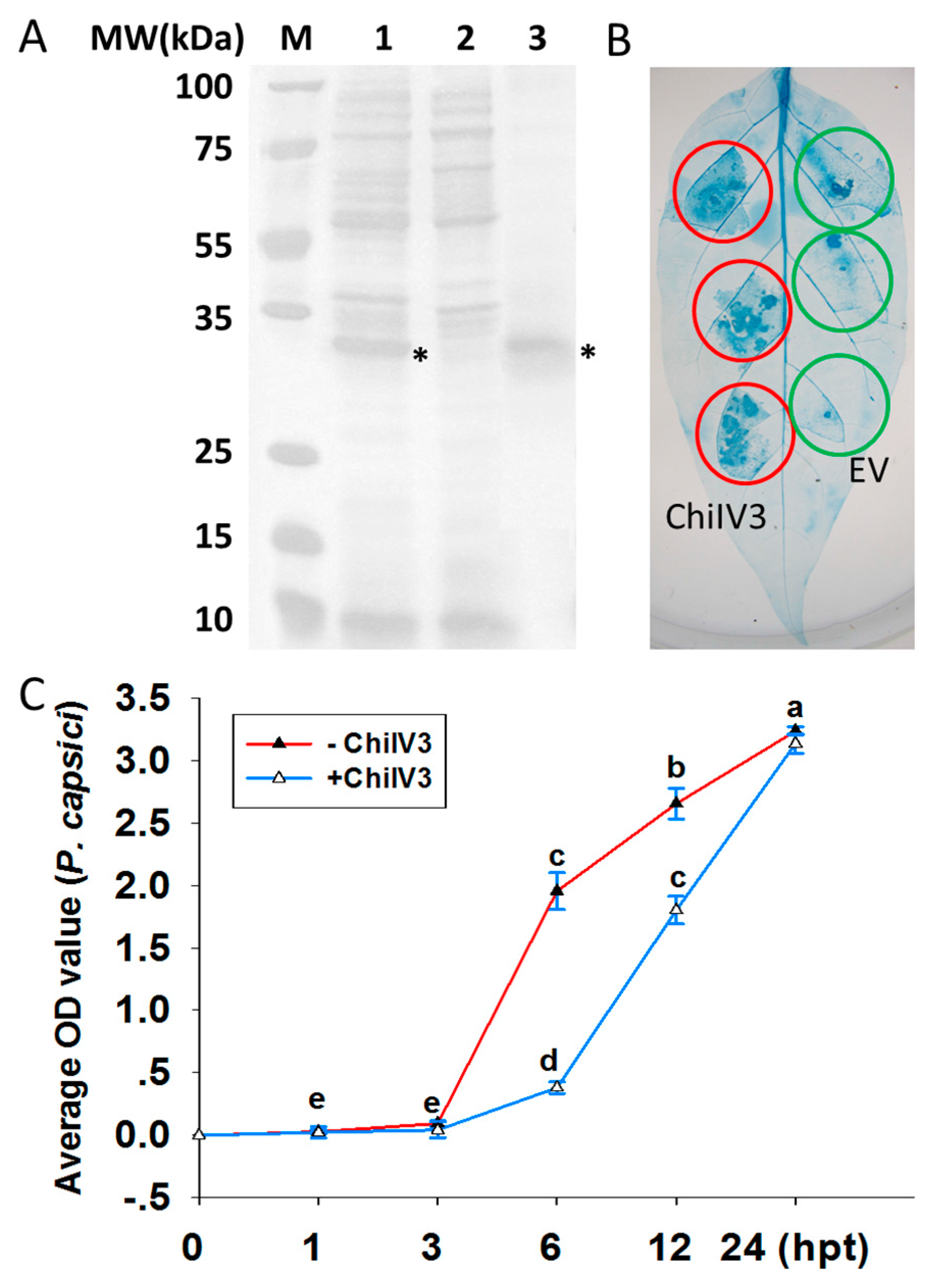
2.4. Prokaryotic Expressed ChiIV3 Inhibited the Growth of P. capsici Mycelia
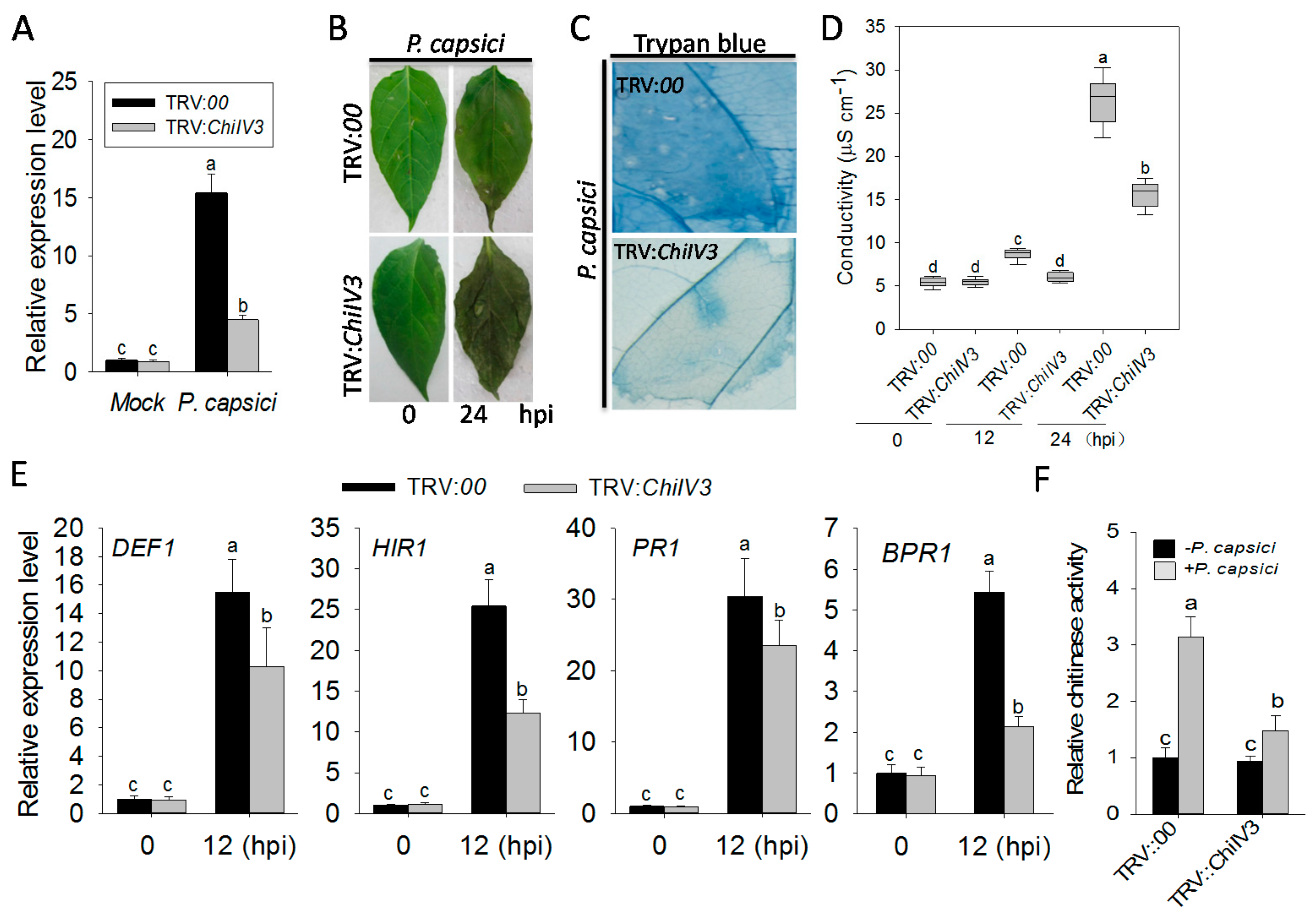
2.5. The Silencing of ChiIV3 Enhanced the Susceptivity of Pepper Plant to P. capsici Inoculation
2.6. Transient Overexpression of ChiIV3 Triggered HR-Like Cell Death and Enhanced the Expression of Immunity Associated Marker Genes
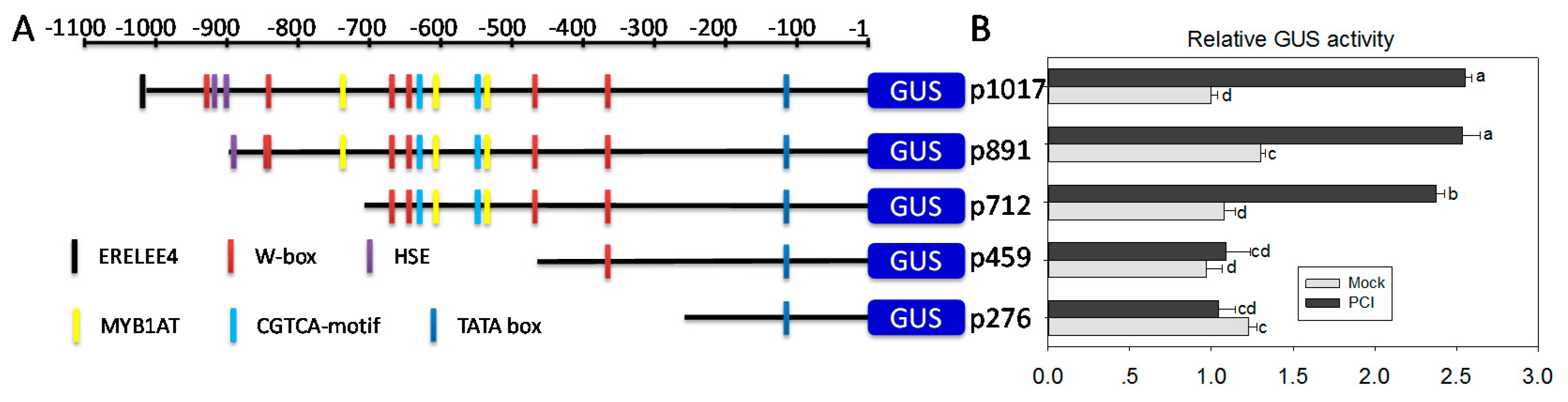
2.7. A Region from −459 bp to −712 bp in the Promoter of ChiIV3 Is Adequate for the PCI-Response of ChiIV3
2.8. W5 Was Found to Be the Only W-Box Conferring the Response of pChiIV3 to PCI
3. Discussion
4. Materials and Methods
4.1. Plant Material and Plant Cultivation
4.2. Pathogen Inoculation
4.3. Isolation of ChiIV3 Promoter
4.4. Amplification of 5′ Deletions or Site-Directed Mutagenesis of pChiIV3 by PCR
4.5. Vector Construction by Gateway Cloning Technique
4.6. Tobacco Transformation
4.7. Prokaryotic Expression of ChiIV3
4.8. Chitinase Activity Measurement
4.9. The Maximal Photochemical Quantum Efficiency of Photosystem II
4.10. Fluorometric Assays for GUS Activity
4.11. Agrobacterium-Mediated Transient Expression Assay
4.12. Western Blotting Assay
4.13. Subcellular Localization
4.14. Total RNA Isolation and Quantitative Real-Time PCR Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Hein, I.; Gilroy, E.M.; Armstrong, M.R.; Birch, P.R. The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 2009, 10, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Katagiri, F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010, 13, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.; Nurnberger, T.; Joosten, M.H. Of pamps and effectors: The blurred pti-eti dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Greeff, C.; Roux, M.; Mundy, J.; Petersen, M. Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 2012, 3, 209. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.S.; Masuta, C. Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr. Opin. Plant Biol. 2014, 20, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.F.; Lin, S.J.; Wang, N.M.; Wu, H.J.; Chen, Y.H. Plant chitinase III Ziz m 1 stimulates multiple cytokines, most predominantly interleukin-13, from peripheral blood mononuclear cells of latex-fruit allergic patients. Ann. Allergy Asthma Immunol. 2012, 108, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Kang, K.K.; Hur, Y.; Lim, Y.P.; Nou, I.S. Molecular characterization of stress resistance-related chitinase genes of Brassica rapa. Plant Physiol. Biochem. 2012, 58, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Itoh, Y.; Takahashi, K.; Takizawa, H.; Nikaidou, N.; Tanaka, H.; Nishihashi, H.; Watanabe, T.; Nishizawa, Y. Family 19 chitinase of Streptomyces griseus HUT6037 increases plant resistance to the fungal disease. Biosci. Biotechnol. Biochem. 2003, 67, 847–855. [Google Scholar] [CrossRef] [PubMed]
- Amian, A.A.; Papenbrock, J.; Jacobsen, H.J.; Hassan, F. Enhancing transgenic pea (Pisum Sativum L.) resistance against fungal diseases through stacking of two antifungal genes (Chitinase and Glucanase). GM Crops 2011, 2, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, O.; Zeller, S.L.; Schmid, B. Competitive performance of transgenic wheat resistant to powdery mildew. PLoS ONE 2011, 6, e28091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Loon, L.C.; Rep, M.; Pieterse, C.M. Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zan, X.L.; Wu, X.F.; Yao, L.; Chen, Y.L.; Jia, S.W.; Zhao, K.J. Identification of fungus-responsive cis-acting element in the promoter of Brassica juncea chitinase gene, BJCHI1. Plant Sci. 2014, 215–216, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Hwang, B.K. Promoter activation of pepper class ii basic chitinase gene, CAChi2, and enhanced bacterial disease resistance and osmotic stress tolerance in the CAChi2-overexpressing Arabidopsis. Planta 2006, 223, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Jiang, W.; Zhang, L.; Zhang, F.; Shen, Q.; Wang, G.; Tang, K. AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua. PLoS ONE 2013, 8, e57657. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Hwang, B.K. Isolation, partial sequencing, and expression of pathogenesis-related cDNA genes from pepper leaves infected by Xanthomonas campestris pv. Vesicatoria. Mol. Plant Microbe Interact. 2000, 13, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Jung, H.W.; Kim, Y.J.; Hwang, B.K. Pepper gene encoding a basic class II chitinase is inducible by pathogen and ethephon. Plant Sci. 2000, 159, 39–49. [Google Scholar] [CrossRef]
- Hong, J.K.; Hwang, B.K. Induction by pathogen, salt and drought of a basic class II chitinase mRNA and its in situ localization in pepper (Capsicum annuum). Physiol. Plant 2002, 114, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Kim, N.H.; Hwang, B.K. The Capsicum annuum class IV chitinase chitiv interacts with receptor-like cytoplasmic protein kinase PIK1 to accelerate PIK1-triggered cell death and defence responses. J. Exp. Bot. 2015, 66, 1987–1999. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Tindwa, H.; Lee, Y.S.; Naing, K.W.; Hong, S.H.; Nam, Y.; Kim, K.Y. Biocontrol of anthracnose in pepper using chitinase, β-1,3 glucanase, and 2-furancarboxaldehyde produced by streptomyces cavourensis SY224. J. Microbiol. Biotechnol. 2012, 22, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.A.; Low, D.; Kamba, A.; Llado, V.; Mizoguchi, E. Oral caffeine administration ameliorates acute colitis by suppressing chitinase 3-like 1 expression in intestinal epithelial cells. J. Gastroenterol. 2014, 49, 1206–1216. [Google Scholar] [PubMed]
- Liu, Z.Q.; Qiu, A.L.; Shi, L.P.; Cai, J.S.; Huang, X.Y.; Yang, S.; Wang, B.; Shen, L.; Huang, M.K.; Mou, S.L.; et al. SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J. Exp. Bot. 2015, 66, 3683–3698. [Google Scholar] [PubMed]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [PubMed]
- Choi, H.W.; Kim, N.H.; Lee, Y.K.; Hwang, B.K. The pepper extracellular xyloglucan-specific endo-β-1,4-glucanase inhibitor protein gene, CaXEGIP1, is required for plant cell death and defense responses. Plant Physiol. 2013, 161, 384–396. [Google Scholar] [PubMed]
- Choi, D.S.; Hwang, I.S.; Hwang, B.K. Requirement of the cytosolic interaction between PATHOGENESIS-RELATED PROTEIN10 and LEUCINE-RICH REPEAT PROTEIN1 for cell death and defense signaling in pepper. Plant Cell 2012, 24, 1675–1690. [Google Scholar] [PubMed]
- Knoth, C.; Eulgem, T. The oomycete response gene LURP1 is required for defense against Hyaloperonospora parasitica in Arabidopsis thaliana. Plant J. 2008, 55, 53–64. [Google Scholar] [PubMed]
- Oliveira, M.B.; Junior, M.L.; Grossi-de-Sa, M.F.; Petrofeza, S. Exogenous application of methyl jasmonate induces a defense response and resistance against Sclerotinia sclerotiorum in dry bean plants. J. Plant Physiol. 2015, 182, 13–22. [Google Scholar] [PubMed]
- Widjaja, I.; Lassowskat, I.; Bethke, G.; Eschen-Lippold, L.; Long, H.H.; Naumann, K.; Dangl, J.L.; Scheel, D.; Lee, J. A protein phosphatase 2C, responsive to the bacterial effector AvrRpm1 but not to the AvrB effector, regulates defense responses in Arabidopsis. Plant J. 2010, 61, 249–258. [Google Scholar] [PubMed]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant. Biol. 2014, 20, 35–46. [Google Scholar] [PubMed]
- Moore, J.W.; Loake, G.J.; Spoel, S.H. Transcription dynamics in plant immunity. Plant Cell 2011, 23, 2809–2820. [Google Scholar] [PubMed]
- Adachi, H.; Nakano, T.; Miyagawa, N.; Ishihama, N.; Yoshioka, M.; Katou, Y.; Yaeno, T.; Shirasu, K.; Yoshioka, H. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 2015, 27, 2645–2663. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Weigman, V.J.; Chang, H.S.; McDowell, J.M.; Holub, E.B.; Glazebrook, J.; Zhu, T.; Dangl, J.L. Gene expression signatures from three genetically separable resistance gene signaling pathways for downy mildew resistance. Plant Physiol. 2004, 135, 1129–1144. [Google Scholar] [CrossRef] [PubMed]
- Curto, M.; Krajinski, F.; Schlereth, A.; Rubiales, D. Transcriptional profiling of Medicago truncatula during Erysiphe pisi infection. Front. Plant Sci. 2015, 6, 517. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Eulgem, T.; Rushton, P.J.; Schmelzer, E.; Hahlbrock, K.; Somssich, I.E. Early nuclear events in plant defence signalling: Rapid gene activation by WRKY transcription factors. EMBO J. 1999, 18, 4689–4699. [Google Scholar] [CrossRef] [PubMed]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Zander, M.; La Camera, S.; Lamotte, O.; Metraux, J.P.; Gatz, C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010, 61, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Fink, J.S.; Verhave, M.; Kasper, S.; Tsukada, T.; Mandel, G.; Goodman, R.H. The CGTCA sequence motif is essential for biological activity of the vasoactive intestinal peptide gene cAMP-regulated enhancer. Proc. Natl. Acad. Sci. USA 1988, 85, 6662–6666. [Google Scholar] [CrossRef] [PubMed]
- Ramonell, K.; Berrocal-Lobo, M.; Koh, S.; Wan, J.; Edwards, H.; Stacey, G.; Somerville, S. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 2005, 138, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Rowland, O.; Ludwig, A.A.; Merrick, C.J.; Baillieul, F.; Tracy, F.E.; Durrant, W.E.; Fritz-Laylin, L.; Nekrasov, V.; Sjolander, K.; Yoshioka, H.; et al. Functional analysis of AVR9/CF-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 2005, 17, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase Nudt7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Veronese, P.; Nakagami, H.; Bluhm, B.; Abuqamar, S.; Chen, X.; Salmeron, J.; Dietrich, R.A.; Hirt, H.; Mengiste, T. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell 2006, 18, 257–273. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, N.; Yoshioka, H. Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 2012, 15, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Garner, C.M.; Gassmann, W. New clues in the nucleus: Transcriptional reprogramming in effector-triggered immunity. Front. Plant Sci. 2013, 4, 364. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.N., Jr.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 1993, 14, 748–750. [Google Scholar] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.W.; Haag, J.R.; Pontes, O.; Opper, K.; Juehne, T.; Song, K.; Pikaard, C.S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006, 45, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.J.; Mendel, R.R.; Schiemann, J.; Simoens, C.; Inze, D. High meiotic stability of a foreign gene introduced into tobacco by Agrobacterium-mediated transformation. Mol. Gen. Genet. 1987, 207, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Wirth, S.J.; Wolf, G.A. Dye-labelled substrates for the assay and detection of chitinase and lysozyme activity. J. Microbiol. Meth. 1990, 12, 197–205. [Google Scholar] [CrossRef]
- Hsu, S.C.; Lockwood, J.L. Powdered chitin agar as a selective medium for enumeration of actinomycetes in water and soil. Appl. Microbiol. 1975, 29, 422–426. [Google Scholar] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Li, P.; Chen, L.; Zhou, Y.; Xia, X.; Shi, K.; Chen, Z.; Yu, J. Brassinosteroids-induced systemic stress tolerance was associated with increased transcripts of several defence-related genes in the phloem in Cucumis sativus. PLoS ONE 2013, 8, e66582. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yang, S.; Yan, Y.; Xiao, Z.; Cheng, J.; Wu, J.; Qiu, A.; Lai, Y.; Mou, S.; Guan, D.; et al. CaWRKY6 transcriptionally activates CaWRKY40, regulates Ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 2015, 66, 3163–3174. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Wang, Y.; She, J.; Lei, Y.; Liu, Z.; Eulgem, T.; Lai, Y.; Lin, J.; Yu, L.; Lei, D.; et al. Overexpression of CaWRKY27, a subgroup IIe WRKY transcription factor of Capsicum annuum, positively regulates tobacco resistance to Ralstonia solanacearum infection. Physiol. Plant 2014, 150, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Kuang, H.; Chen, C.; Yan, J.; Do-Umehara, H.C.; Liu, X.Y.; Dada, L.; Ridge, K.M.; Chandel, N.S.; Liu, J. The kinase Jnk2 promotes stress-induced mitophagy by targeting the small mitochondrial form of the tumor suppressor ARF for degradation. Nat. Immunol. 2015, 16, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Shi, L.; Yang, S.; Lin, Y.; Weng, Y.; Li, X.; Hussain, A.; Noman, A.; He, S. Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection. Int. J. Mol. Sci. 2017, 18, 1661. https://doi.org/10.3390/ijms18081661
Liu Z, Shi L, Yang S, Lin Y, Weng Y, Li X, Hussain A, Noman A, He S. Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection. International Journal of Molecular Sciences. 2017; 18(8):1661. https://doi.org/10.3390/ijms18081661
Chicago/Turabian StyleLiu, Zhiqin, Lanping Shi, Sheng Yang, Youquan Lin, Yahong Weng, Xia Li, Ansar Hussain, Ali Noman, and Shuilin He. 2017. "Functional and Promoter Analysis of ChiIV3, a Chitinase of Pepper Plant, in Response to Phytophthora capsici Infection" International Journal of Molecular Sciences 18, no. 8: 1661. https://doi.org/10.3390/ijms18081661





