Spatial Distribution Patterns of Root-Associated Bacterial Communities Mediated by Root Exudates in Different Aged Ratooning Tea Monoculture Systems
Abstract
:1. Introduction
2. Results
2.1. The Problems of Camellia sinensis (L.) Plantation under Continuous Monoculture
2.2. Soil Physio-Chemical Properties and Root Exudates
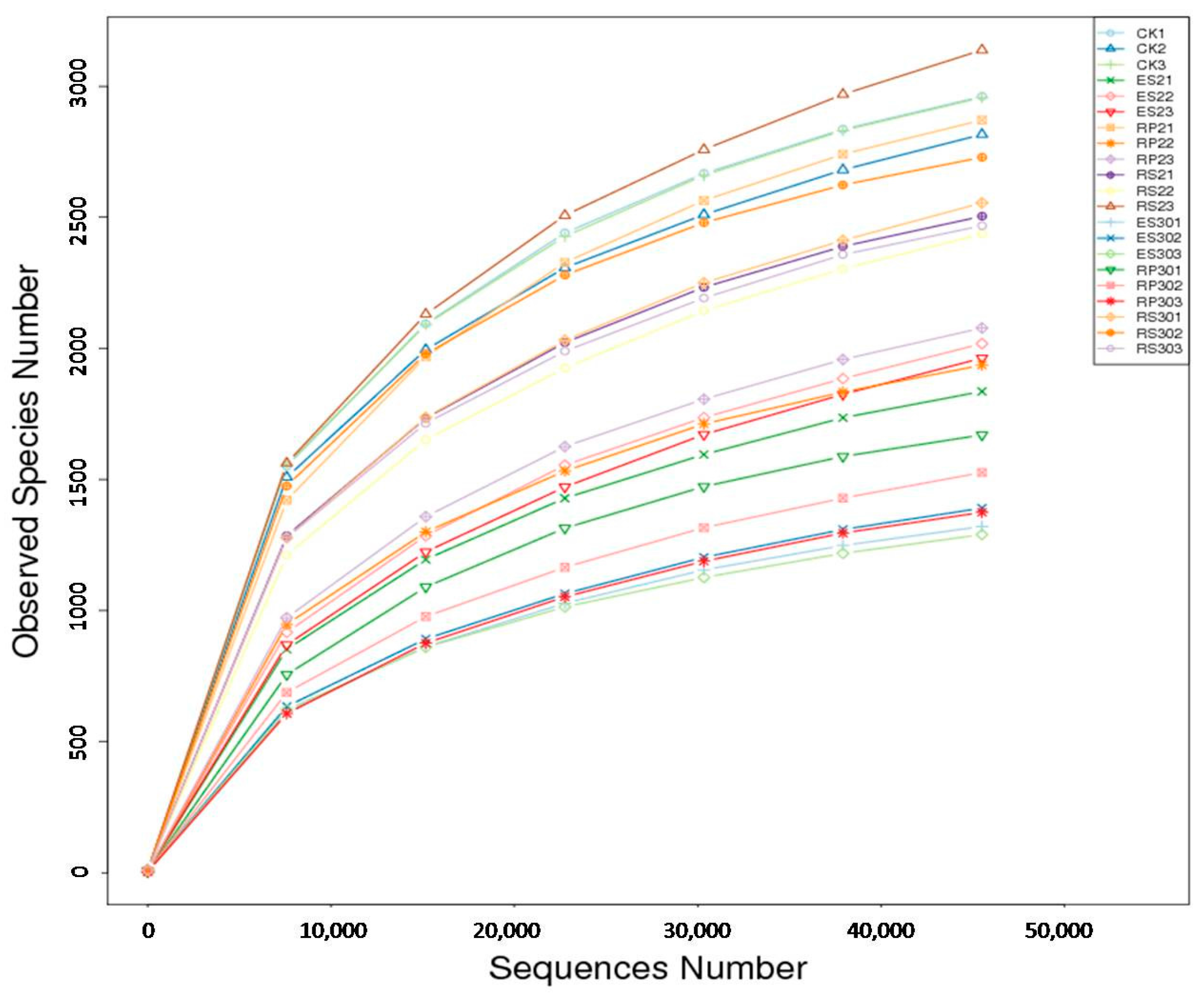
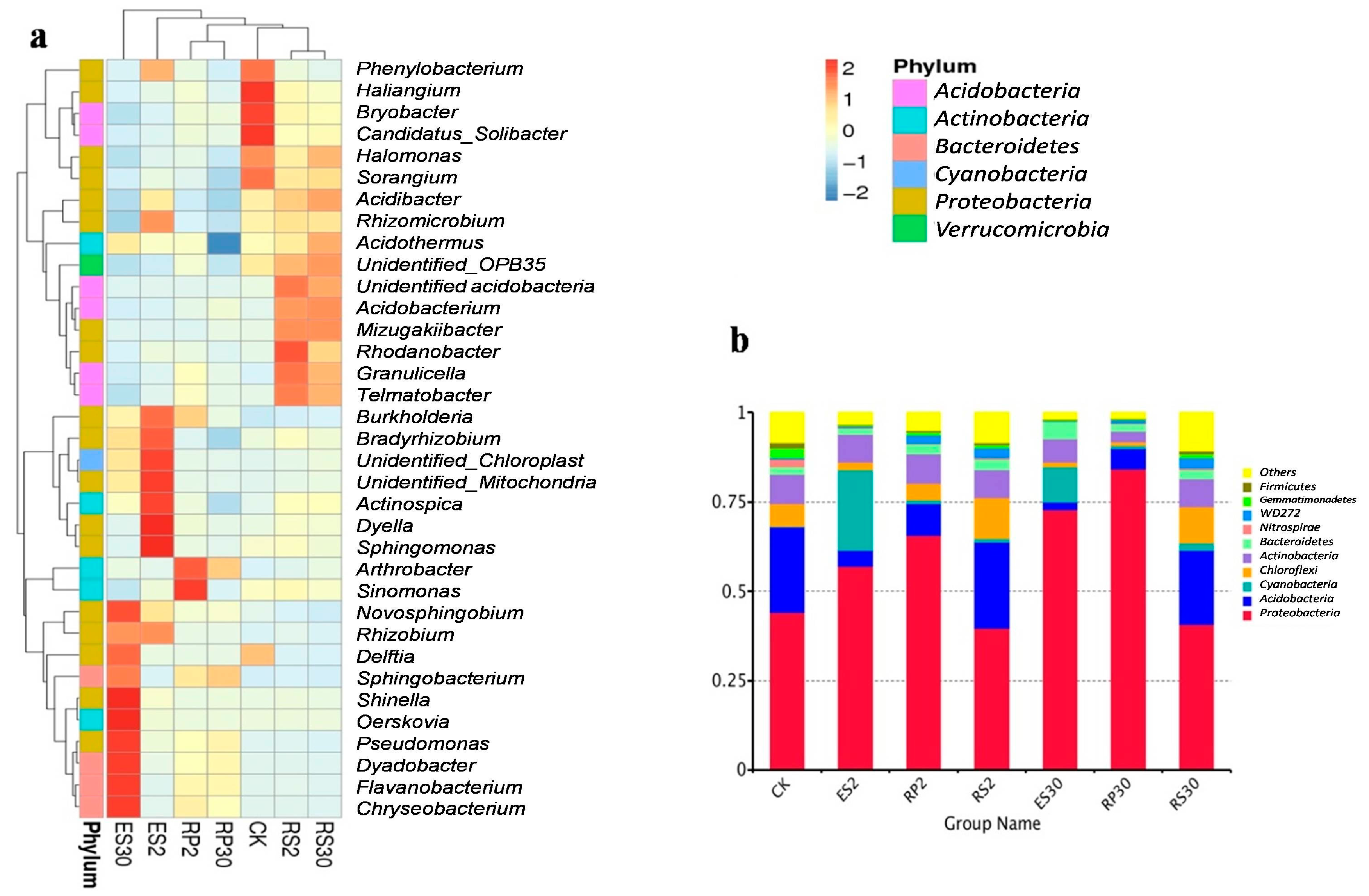
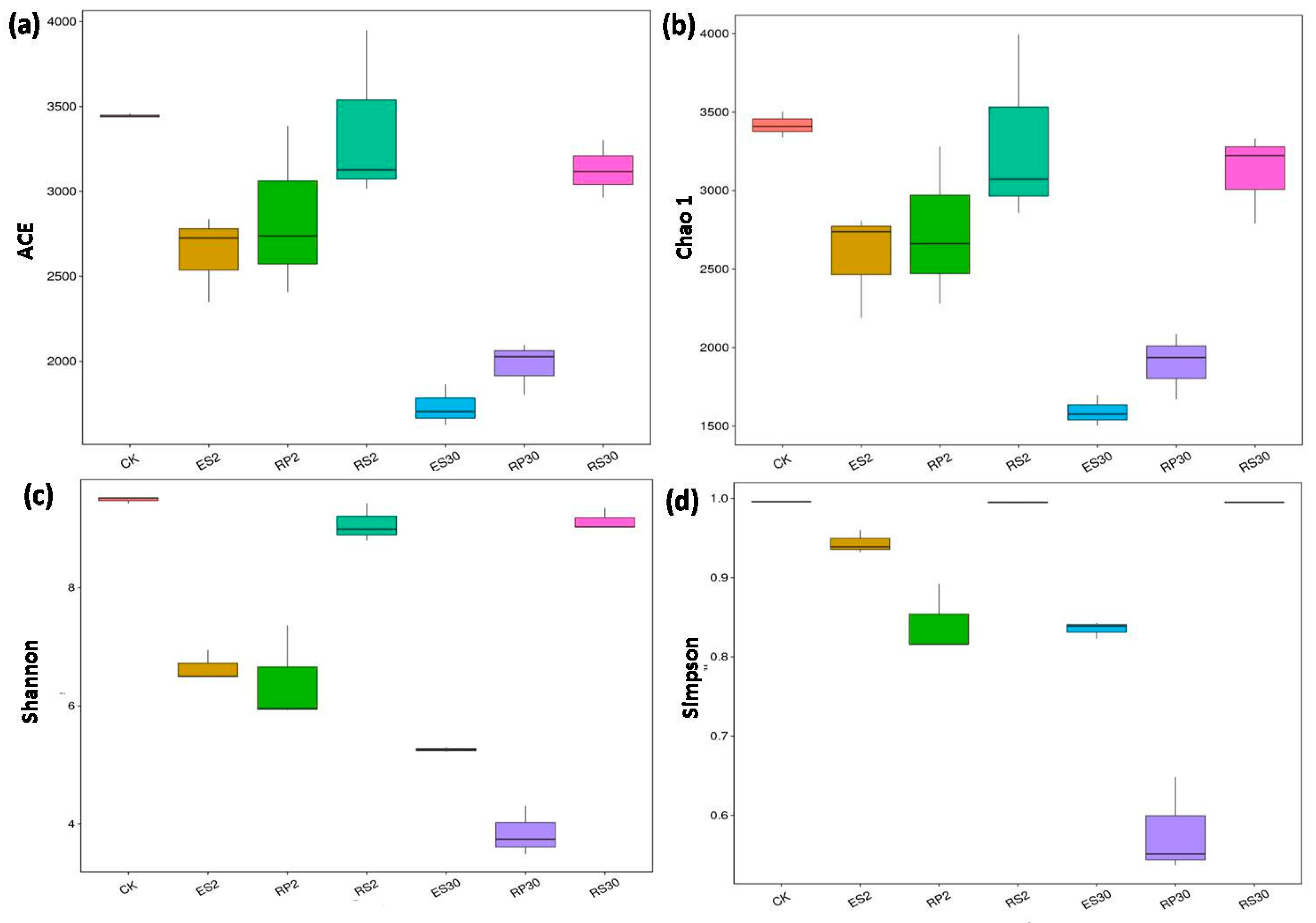
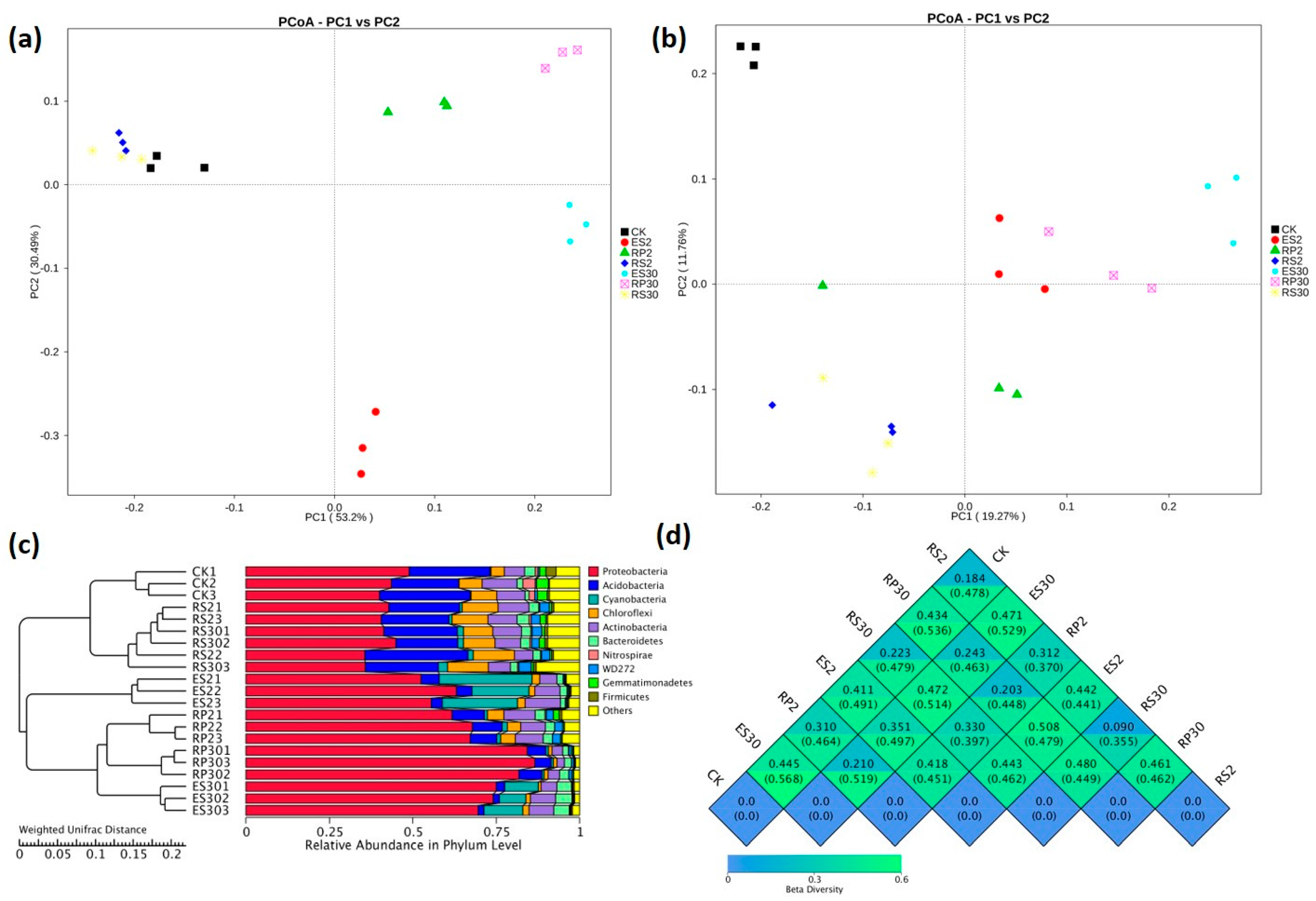
2.3. 16S rDNA-Based Meta-Genomic Analysis of Tea Root-Associated Bacteria
2.4. Distinct and Overlapping Bacterial Communities of Root-Associated Bacterial Communities across Different Tea Plantations of Different Ages
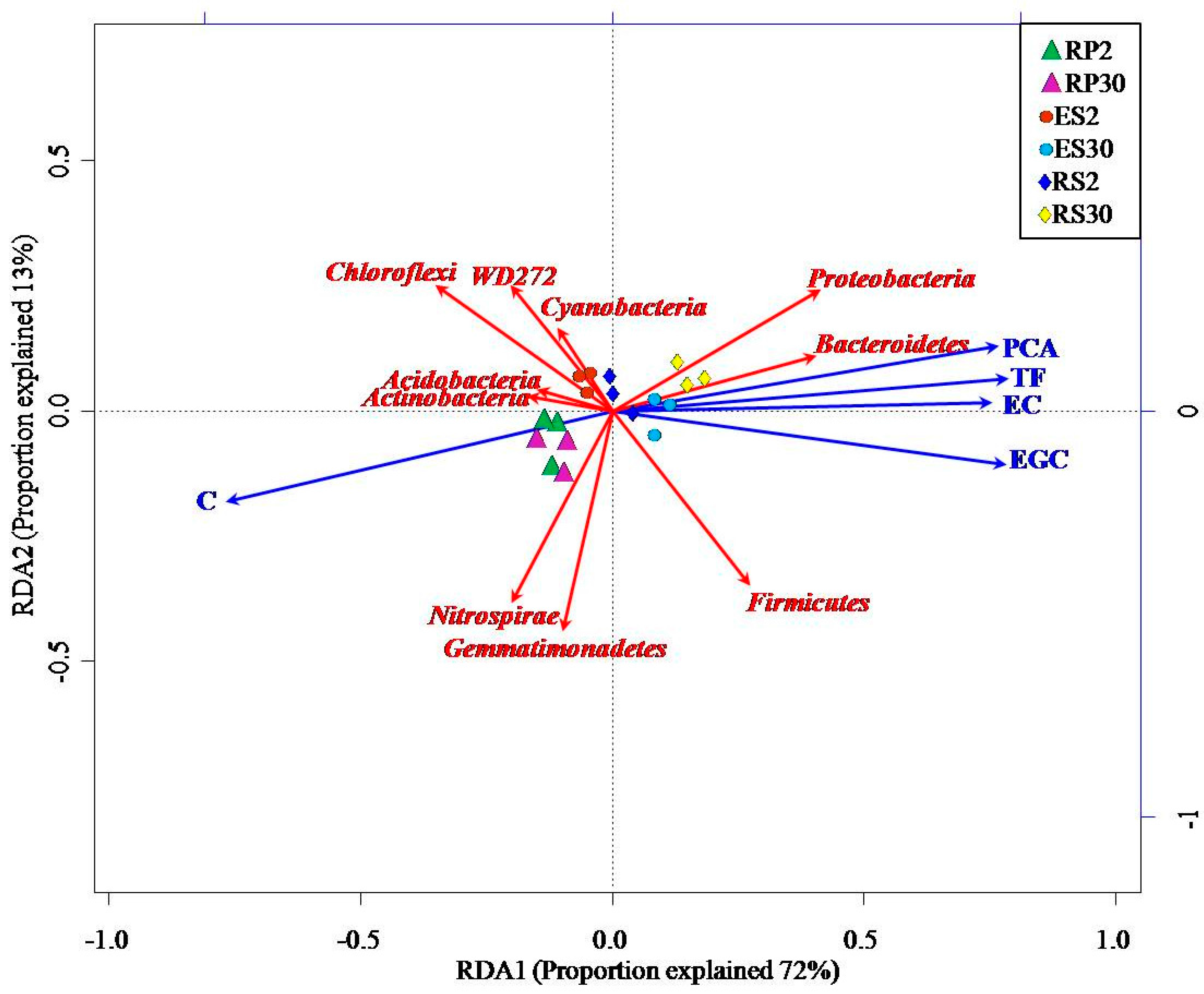
2.5. Interactions of Bacterial Abundance with Soil Physio-Chemical Properties and Root Exudates
3. Discussion
4. Materials and Methods
4.1. The Collection of Plant and Rhizosphere Soil Samples
4.2. Quality Parameters and Soil Properties Determination
4.3. Identification and Quantification of Allelochemicals from Tea Root Exudates
4.4. DNA Extraction and Purification
4.5. The Metagenomic Analysis of the Root-Associated Rhizo-Compartment Bacteria
4.6. Statistical and Bioinformatics Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bever, J.D.; Westover, K.M.; Antonovics, J. Incorporating the soil community into plant population dynamics: The utility of the feedback approach. J. Ecol. 1997, 85, 561–573. [Google Scholar] [CrossRef]
- Bever, J.D. Soil community feedback and the coexistence of competitors: Conceptual frameworks and empirical tests. New Phytol. 2003, 157, 465–473. [Google Scholar] [CrossRef]
- Kulmatiski, A.; Beard, K.H.; Stevens, J.R.; Cobbold, S.M. Plantntitors: Conceptual fra-analytical review. Ecol. Lett. 2008, 11, 980–992. [Google Scholar] [CrossRef] [PubMed]
- Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomos, J.N.; Kulmatiski, A.; Schweitzer, J.A. Plant–soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Baxendale, C.; Orwin, K.H.; Poly, F.; Pommier, T.; Bardgett, R.D. Are plant–soil feedback responses explained by plant traits? New Phytol. 2014, 204, 408–423. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.F.; Song, L.X.; Xia, X.J.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Plant-soil feedbacks and soil sickness: From mechanisms to application in agriculture. J. Chem. Ecol. 2013, 39, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Klironomos, J.N. Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 2002, 417, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Van-der-Putten, W.H. Plant defense belowground and spatiotemporal processes in natural vegetation. Ecology 2003, 84, 2269–2280. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; van-der-Putten, W.H.; Wall, D.H. Ecological linkages between aboveground and belowground biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Harrison, K.A.; Bardgett, R.D. Influence of plant species and soil conditions on plant–soil feedback in mixed grassland communities. J. Ecol. 2010, 98, 384–395. [Google Scholar] [CrossRef]
- Muller, C.H. The role of chemical inhibition (allelopathy) in vegetational composition. Bull. Torrey Bot. Club 1966, 93, 332–351. [Google Scholar] [CrossRef]
- Zhao, Y.P.; Lin, S.; Chu, L.; Gao, J.T.; Azeem, S.; Lin, W. Insight into structure dynamics of soil microbiota mediated by the richness of replanted Pseudostellaria heterophylla. Sci. Rep. 2016, 6, 26175. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, L.; Chu, L.; Yang, Y.; Li, Z.; Azeem, S.; Zhang, Z.; Fang, C.; Lin, W. Interaction of Pseudostellaria heterophylla with Fusarium oxysporum f. Sp. Heterophylla mediated by its root exudates in a consecutive monoculture system. Sci. Rep. 2015, 5, 8197. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Wang, J.; Wu, Y.; Lin, S.; Khan, M.U.; Zhang, Z.; Lin, W. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 2016, 6, 26601. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.J.; Bending, G.D.; Chandler, D.; Hilton, S.; Mills, P. Meeting the demand for crop production: The challenge of yield decline in crops grown in short rotations. Biol. Rev. 2012, 87, 52–71. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, H.; Zhang, Z.; Lin, R.; Zhang, Z.; Lin, W. Comparative metaproteomic analysis on consecutively Rehmannia glutinosa-monocultured rhizosphere soil. PLoS ONE 2011, 6, e20611. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, R.; Weinhold, A.; Goldberg, J.; Oh, Y.; Baldwin, I.T. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc. Natl. Acad. Sci. USA 2015, 112, E5013–E5020. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.H.; Wang, J.G. Effect of amendments with different phenolic acids on soil microbial biomass, activity, and community diversity. Appl. Soil Ecol. 2008, 39, 172–179. [Google Scholar] [CrossRef]
- Utkhede, R.S. Soil sickness, replant problem or replant disease and its integrated control. Allelopathy J. 2006, 18, 23–38. [Google Scholar]
- Li, C.; Li, X.; Kong, W.; Wu, Y.; Wang, J. Effect of monoculture soybean on soil microbial community in the northeast China. Plant Soil 2010, 330, 423–433. [Google Scholar] [CrossRef]
- Liua, X.; Herbert, S.J. Fifteen years of research examining cultivation of continuous soybean in northeast china: A review. Field Crops Res. 2002, 79, 1–7. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Arafat, Y.; Lin, W.; Jiang, Y.; Weng, B.; Lin, W. Characterizing rhizosphere microbial communities in long-term monoculture tea orchards by fatty acid profiles and substrate utilization. Eur. J. Soil Biol. 2017, 81, 48–54. [Google Scholar] [CrossRef]
- Ferrazzano, G.F.; Roberto, L.; Amato, I.; Cantile, T.; Sangianantoni, G.; Ingenito, A. Antimicrobial properties of green tea extract against cariogenic microflora: An in vivo study. J. Med. Food 2011, 14, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Sakanaka, S.; Juneja, L.R.; Taniguchi, M. Antimicrobial effects of green tea polyphenols on thermophilic spore-forming bacteria. J. Biosci. Bioeng. 2000, 90, 81–85. [Google Scholar] [CrossRef]
- Taylor, P.W.; Hamilton-Miller, J.M.T.; Stapleton, P.D. Antimicrobial properties of green tea catechins. Food Sci. Technol. Bull. 2005, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Veluri, R.; Weir, T.L.; Bais, H.P.; Stermitz, F.R.; Vivanco, J.M. Phytotoxic and antimicrobial activities of catechin derivatives. J. Agric. Food Chem. 2004, 52, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Henning, S.M.; Fajardo-Lira, C.; Lee, H.W.; Youssefian, A.A.; Go, V.L.W.; Heber, D. Catechin content of 18 teas and a green tea extract supplement correlates with the antioxidant capacity. Nutr. Cancer 2003, 45, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Carando, S.; Teissedre, P.L. Catechin and procyanidin levels in french wines: Contribution to dietary intake. Basic Life Sci. 1999, 66, 725–737. [Google Scholar] [PubMed]
- Chunmei, D.; Jiabo, W.; Weijun, K.; Cheng, P.; Xiaohe, X. Investigation of anti-microbial activity of catechin on Escherichia coli growth by microcalorimetry. Environ. Toxicol. Pharmacol. 2010, 30, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Walker, T.S.; Kennan, A.J.; Stermitz, F.R.; Vivanco, J.M. Structure-dependent phytotoxicity of catechins and other flavonoids: Flavonoid conversions by cell-free protein extracts of Centaurea maculosa (spotted knapweed) roots. J. Agric. Food Chem. 2003, 51, 897–901. [Google Scholar] [CrossRef] [PubMed]
- Callaway, R.M.; Thelen, G.C.; Rodriguez, A.; Holben, W.E. Soil biota and exotic plant invasion. Nature 2004, 427, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.M.; Li, T.C.; Jhan, Y.L.; Weng, J.H.; Chou, C.H. The impact of microbial biotransformation of catechin in enhancing the allelopathic effects of Rhododendron formosanum. PLoS ONE 2013, 8, e85162. [Google Scholar] [CrossRef] [PubMed]
- Perry, L.G.; Thelen, G.C.; Ridenour, W.M.; Callaway, R.M.; Paschke, M.W.; Vivanco, J.M. Concentrations of the allelochemical (±)-catechin in Centaurea maculosa soils. J. Chem. Ecol. 2007, 33, 2337–2344. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Kemmitt, S.J.; Brookes, P.C. Soil microbial biomass and activity in Chinese tea gardens of varying stand age and productivity. Soil Biol. Biochem. 2007, 39, 1468–1478. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, Z.; Li, Z.W.; Jiang, Y.H.; Weng, B.Q.; Lin, W.X. Variations of rhizosphere bacterial communities in tea (Camellia sinensis L.) continuous cropping soil by high-throughput pyrosequencing approach. J. Appl. Microbiol. 2016, 121, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Liu, C.; Li, D. Effects of different autotoxins on antioxidant enzymes and chemical compounds in tea (Camellia sinensis L.) kuntze. Afr. J. Biotechnol. 2011, 10, 7480–7486. [Google Scholar]
- Zhang, X.L.; Pan, Z.G.; Zhou, X.F.; Ni, W.Z. Autotoxicity and continuous cropping obstacles: A review. Chin. J. Soil Sci. 2007, 4, 33. [Google Scholar]
- Pollock, J.L.; Callaway, R.M.; Holben, W. Phytotoxic effects of (±)-catechin in vitro, in soil, and in the field. PLoS ONE 2008, 3, e2536. [Google Scholar]
- Van-der-Putten, W.H. Impacts of soil microbial communities on exotic plant invasions. Trends Ecol. Evol. 2010, 25, 512–519. [Google Scholar]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Buckley, D.H.; Schmidt, T.M. The structure of microbial communities in soil and the lasting impact of cultivation. Microb. Ecol. 2001, 42, 11–21. [Google Scholar] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Li, J.H.; Friedman, C.R.; Wang, H.F. Variation of soil bacterial communities in a chronosequence of rubber tree (Hevea brasiliensis) plantations. Front. Plant Sci. 2017, 8, 849. [Google Scholar] [CrossRef] [PubMed]
- Lynn, T.M.; Liu, Q.; Hu, Y.; Yuan, H.; Wu, X.; Khai, A.A.; Wu, J.; Ge, T. Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch. Microbiol. 2017, 199, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.G. Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace Elem. Med. Biol. 2005, 18, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L. Organic acids in the rhizosphere–a critical review. Plant Soil 1998, 205, 25–44. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Song, X.; Shi, X.; Li, J.; Ye, C. An improved hplc method for simultaneous determination of phenolic compounds, purine alkaloids and theanine in Camellia species. J. Food Compos. Anal. 2008, 21, 559–563. [Google Scholar] [CrossRef]
- Bao, S.D. Soil and Agricultural Chemistry Analysis; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Pansu, M.; Gautheyrou, J. Handbook of Soil Analysis: Mineralogical, Organic and Inorganic Methods; Springer Science & Business Media: Berlin, Germany, 2007. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. Qiime allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Saqib, H.S.A.; You, M.; Gurr, G.M. Multivariate ordination identifies vegetation types associated with spider conservation in brassica crops. PeerJ Prepr. 2017, 5, e3003v3001. [Google Scholar]
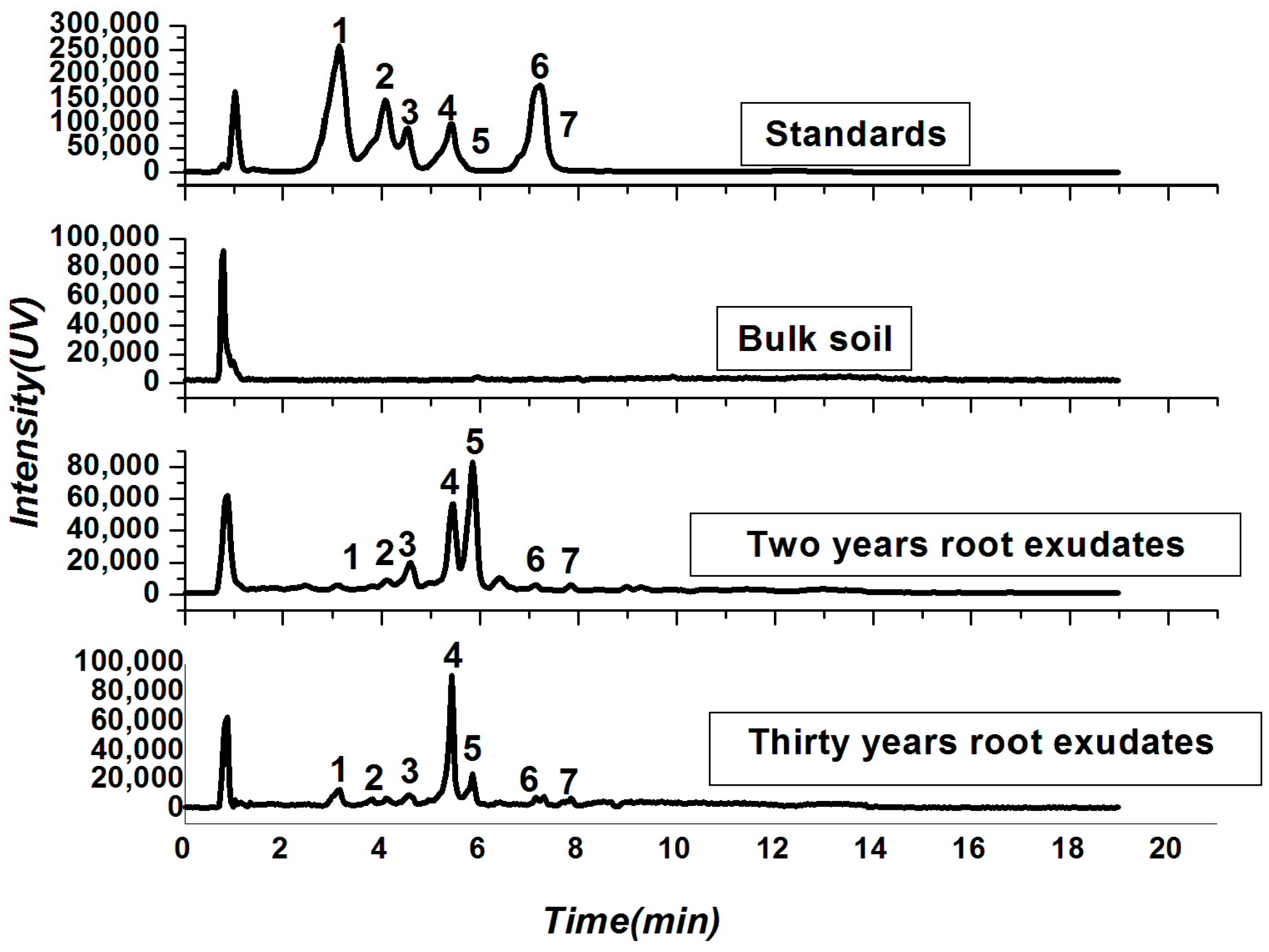

| Treatments | TNN (g/kg) | TPY (mg/kg) | TPP (g/kg) | TAA (g/kg) | WC (%) |
|---|---|---|---|---|---|
| 2YTL | 1.70 a | 0.12 a | 106.40 a | 12.44 a | 0.76 a |
| 30YTL | 0.47 b | 0.02 b | 30.66 b | 8.87 b | 0.56 a |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arafat, Y.; Wei, X.; Jiang, Y.; Chen, T.; Saqib, H.S.A.; Lin, S.; Lin, W. Spatial Distribution Patterns of Root-Associated Bacterial Communities Mediated by Root Exudates in Different Aged Ratooning Tea Monoculture Systems. Int. J. Mol. Sci. 2017, 18, 1727. https://doi.org/10.3390/ijms18081727
Arafat Y, Wei X, Jiang Y, Chen T, Saqib HSA, Lin S, Lin W. Spatial Distribution Patterns of Root-Associated Bacterial Communities Mediated by Root Exudates in Different Aged Ratooning Tea Monoculture Systems. International Journal of Molecular Sciences. 2017; 18(8):1727. https://doi.org/10.3390/ijms18081727
Chicago/Turabian StyleArafat, Yasir, Xiaoya Wei, Yuhang Jiang, Ting Chen, Hafiz Sohaib Ahmed Saqib, Sheng Lin, and Wenxiong Lin. 2017. "Spatial Distribution Patterns of Root-Associated Bacterial Communities Mediated by Root Exudates in Different Aged Ratooning Tea Monoculture Systems" International Journal of Molecular Sciences 18, no. 8: 1727. https://doi.org/10.3390/ijms18081727