Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.)
Abstract
:1. Introduction
2. Results
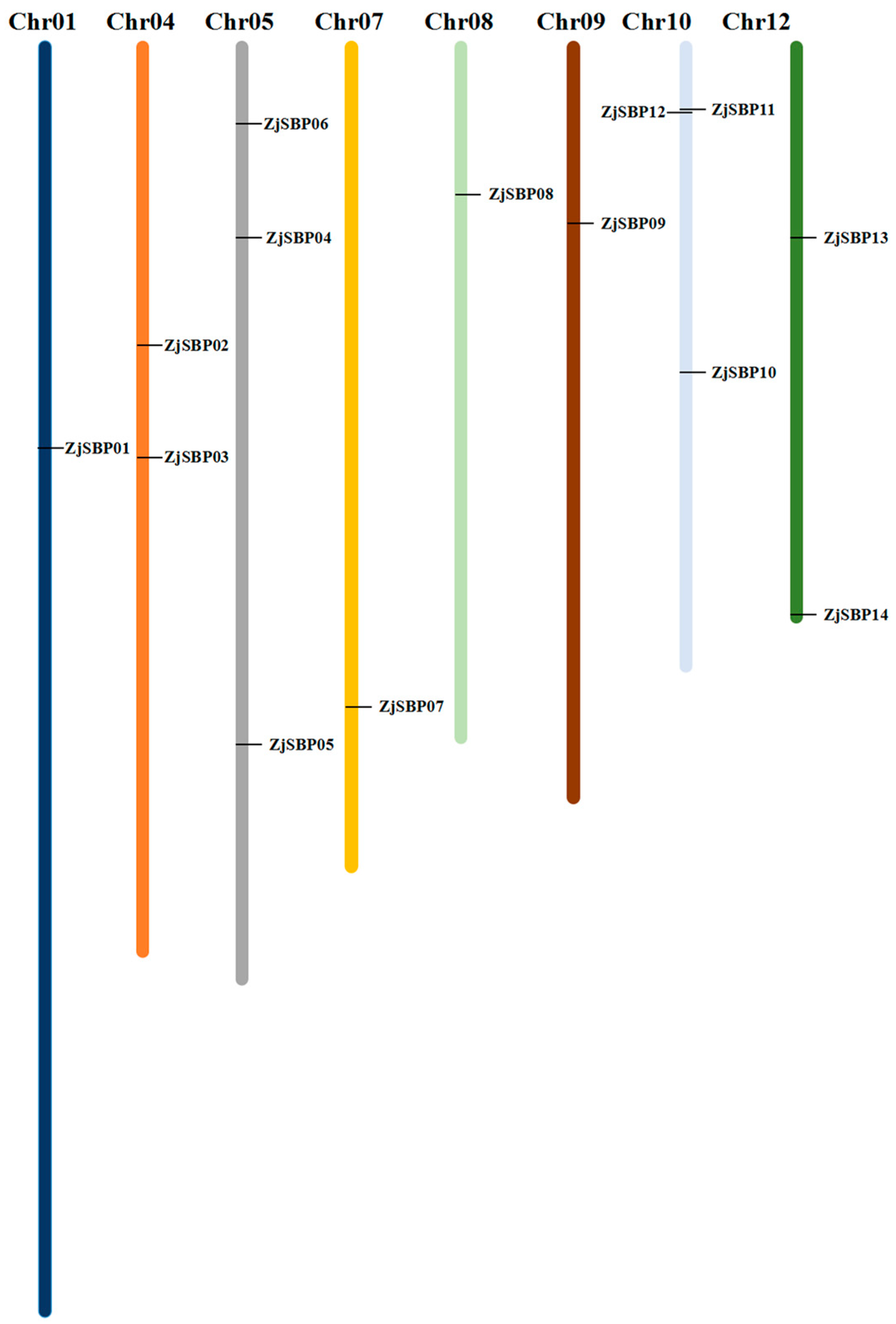
2.1. Identification of the SBP-Box Gene Family in Dongzao
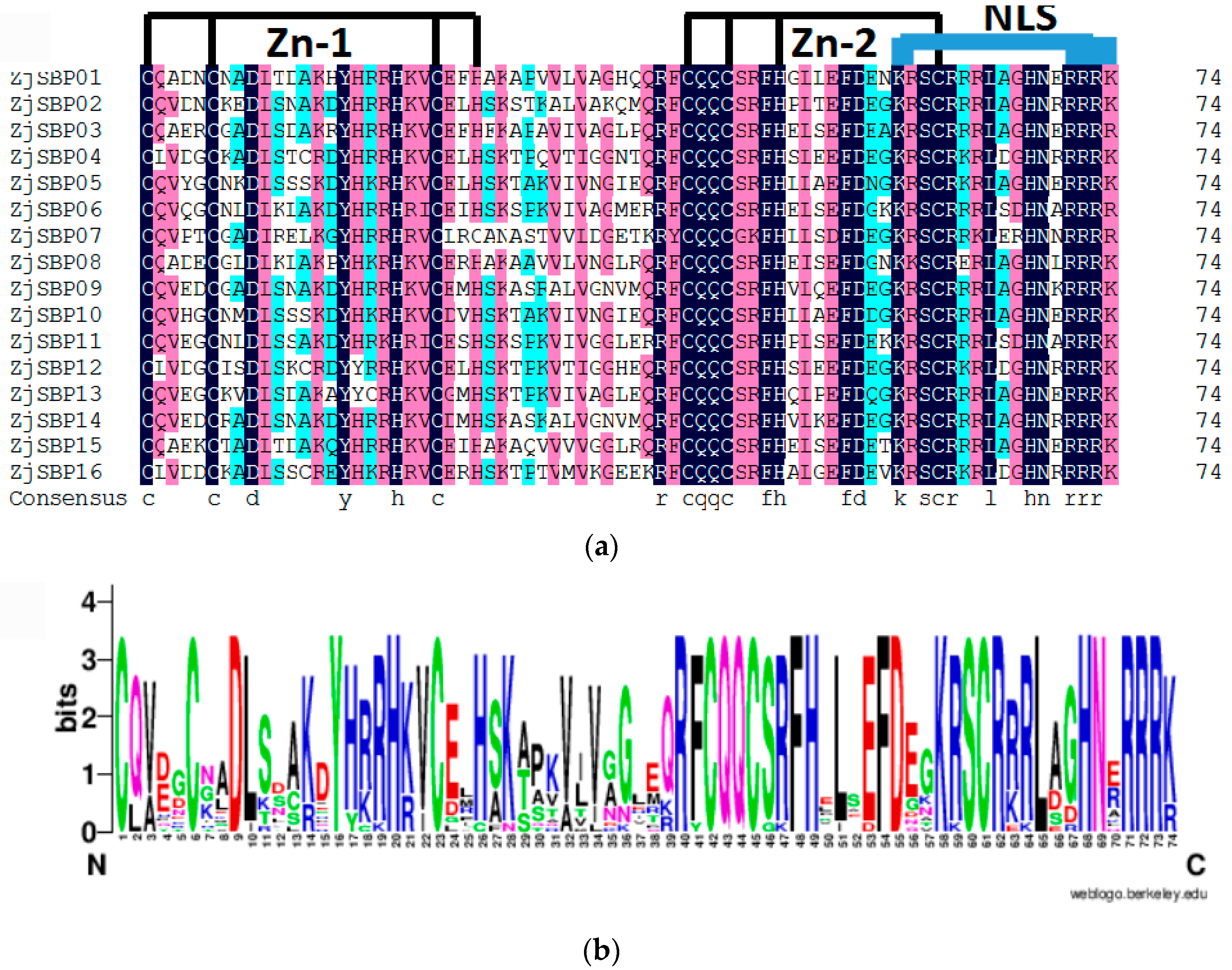
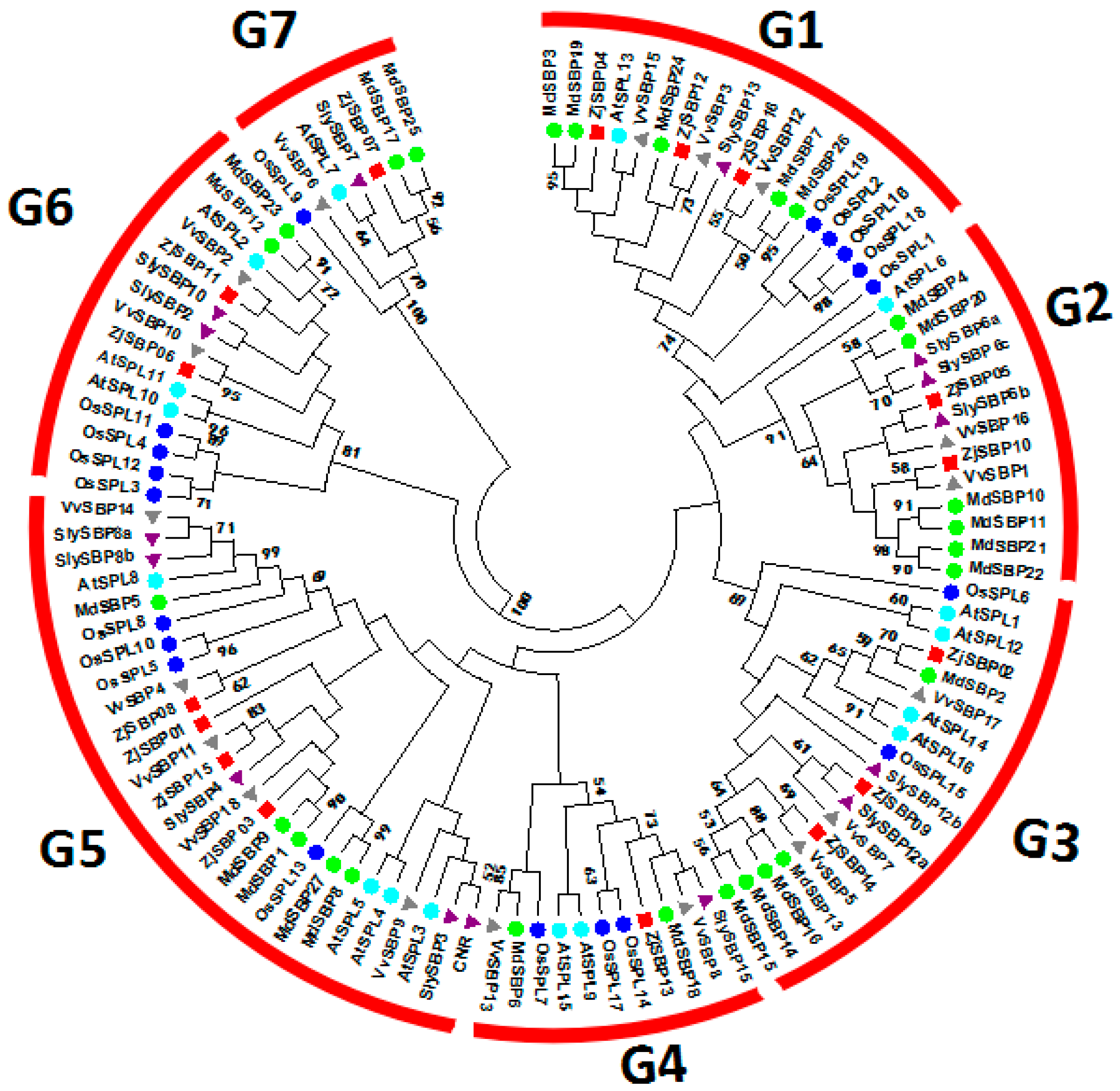
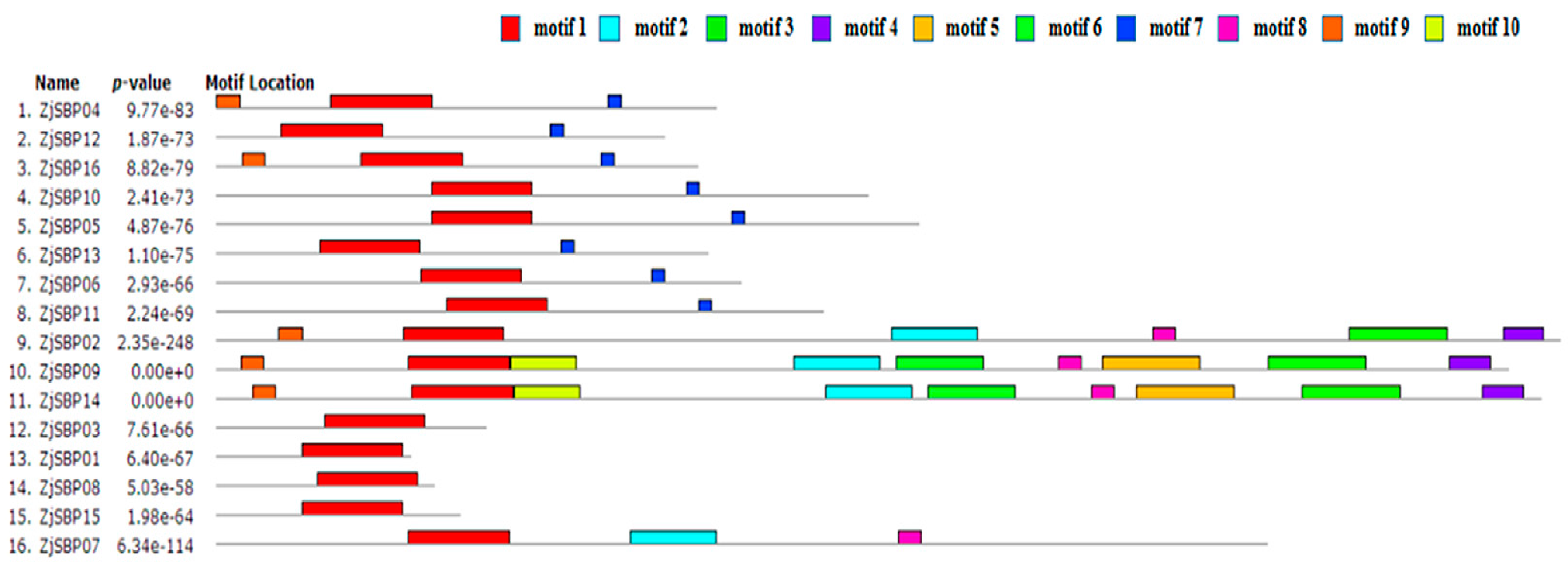
2.2. Sequence Alignments and Phylogenetic Analyses
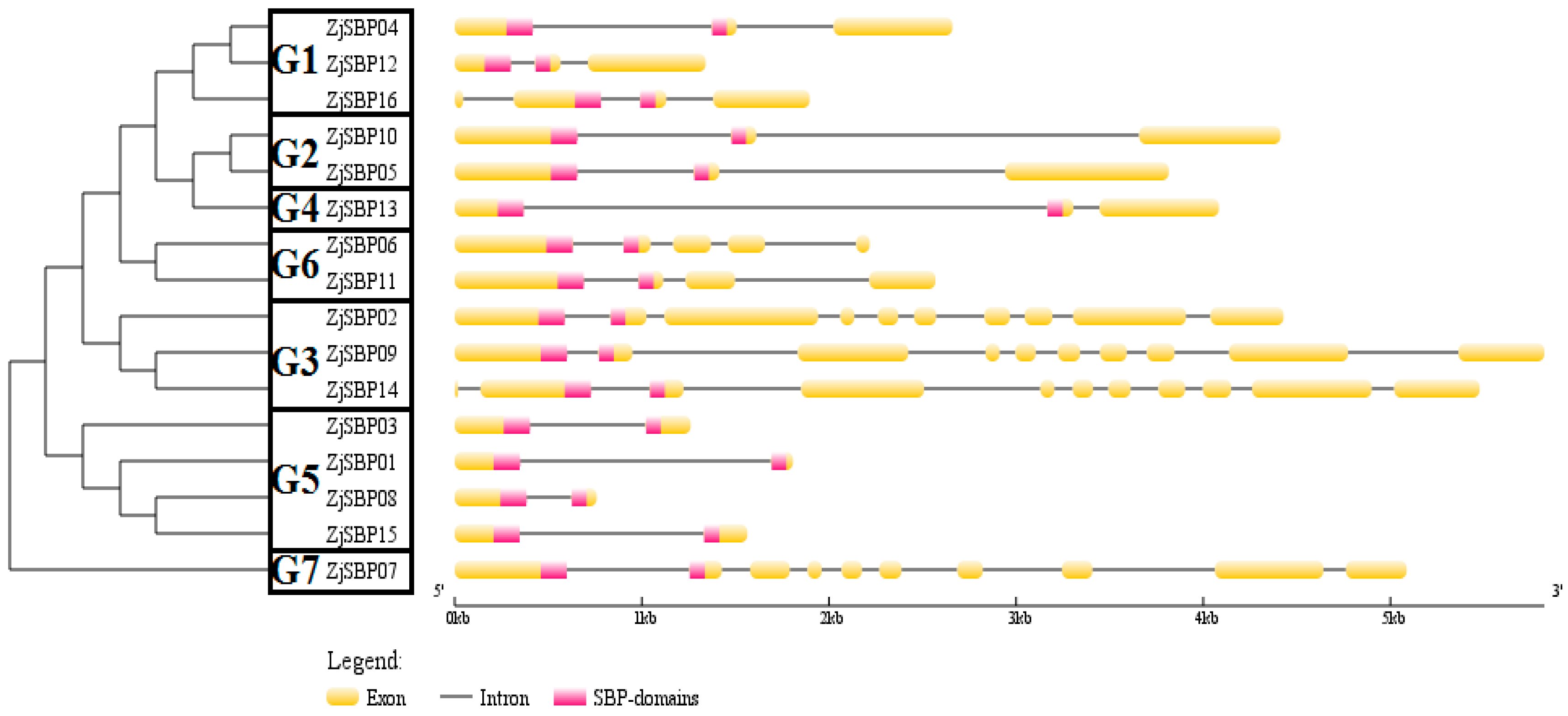
2.3. Gene Structures of the ZjSBP Genes
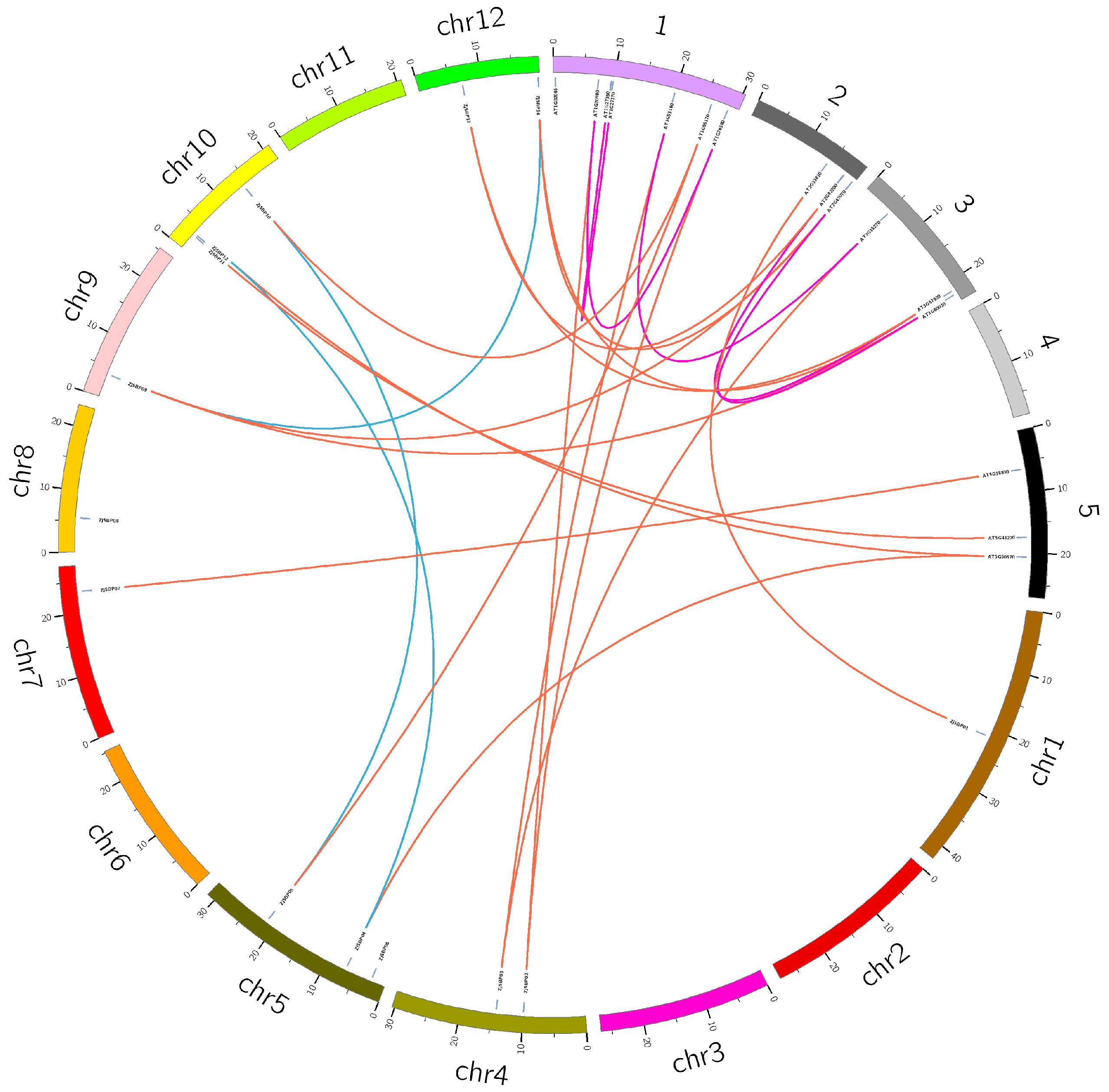
2.4. Homology Analysis
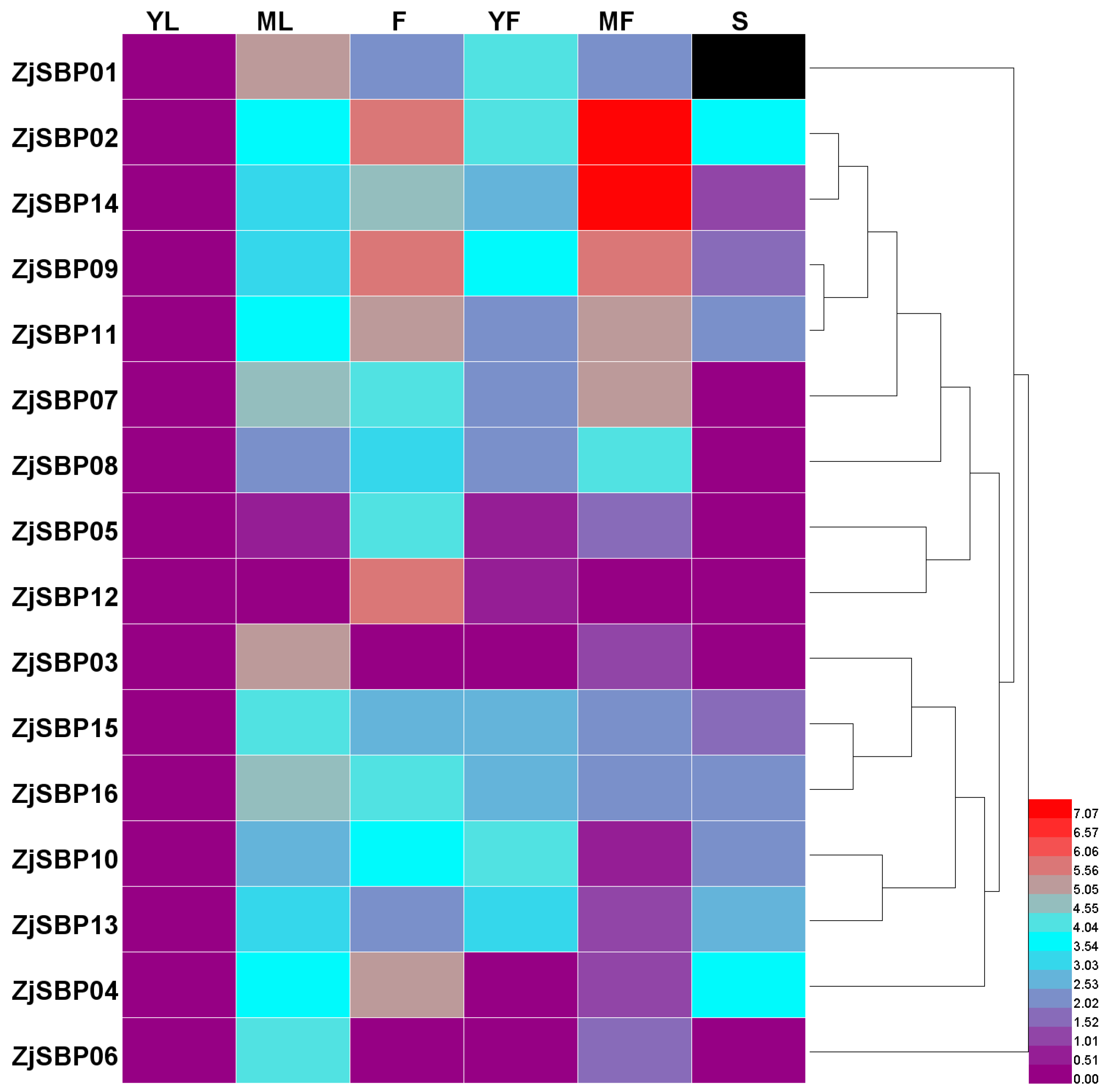
2.5. Expression Analysis of the ZjSBP Genes
2.6. Expression Patterns of the Transcriptome of Different Tissues
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Identification of SBP-Box Genes in Dongzao
4.3. Sequence Alignments, Phylogenetic Analyses and Exon-Intron Structure Determination
4.4. Homology Analysis
4.5. Expression Analysis of the ZjSBP Genes
4.6. Expression Quantity of the Transcriptome of Different Tissues
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus, floral meristem identity gene SQUAMOSA. Mol. Genet. Genom. 1996, 250, 7–16. [Google Scholar]
- Rainer, P.B.; Guido, J.; Heinz, S.; Peter, H. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar]
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Iida, K.; Seki, M.; Sakurai, T.; Satou, M.; Akiyama, K.; Toyoda, T.; Konagaya, A.; Shinozakiet, K. RARTF: Database and tools for complete sets of Arabidopsis transcription factors. DNA Res. 2005, 12, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Höhmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterization of the Arabidopsis SBP-box genes. Gene. 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Xie, K. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X.P. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus×domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.H.; Höhmann, S.; Nettesheim, K.; Saedler, H.; Huijser, P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, P.J.; Kang, S.K.; Park, C.M. MiR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5, genes in Arabidopsis, developmental transitions. Plant Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Höhmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Andrew, J.; Mahmut, T.; Cornelius, S.; Julia, V.; Caroline, O.; Michael, C.J.; James, J.G.; Donald, G.; Graham, B.S. Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 1999, 120, 383–389. [Google Scholar]
- Emma, E.M.; Bovy, A.; Manning, K.; Harrison, L.; Andrews, J.; de Silva, J.; Tucker, G.A.; Seymour, G.B. Effect of the colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004, 136, 4184–4197. [Google Scholar]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Fan, L.P.; Ding, S.D.; Ding, X.L. Nutritional composition of five cultivars of Chinese jujube. Food Chem. 2007, 103, 454–460. [Google Scholar] [CrossRef]
- Liu, M.J. The present status, problems and countermeasures of Chinese jujube production (in Chinese). Rev. China Agric. Sci. Technol. 2000, 2, 76–80. [Google Scholar]
- Huang, J.; Zhang, C.; Zhao, X.; Fei, Z.; Wan, K.; Zhang, Z.; Pang, X.; Yin, X.; Bai, Y.; Sun, X.; et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Zhao, J.; Liu, M.; Liu, P.; Dai, L.; Zhao, Z. Genome-wide characterization of Simple Sequence Repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS ONE 2015, 10, e0127812. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Wu, C.; Wang, M. The jujube (Ziziphus jujuba Mill.) fruit: A review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 2013, 61, 3351–3363. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Wang, Y. China fruit’s monograph—Chinese jujube volume. Beijing China For. Publ. House 1993, 56, 229. [Google Scholar]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315–5326. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Riese, M.; Höhmann, S.; Saedler, H.; Münster, T.; Huijser, P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.C.; Asahina, M.; Liu, P.P.; Kristof, J.R.; Coppersmith, J.L.; Pluskota, W.E.; Bassel, G.W.; Goloviznina, N.A.; Nguyen, T.T.; Martine-Andújar, C.; et al. The regulation of post-germinative transition from the cotyledon-to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis. Seed Sci. Res. 2010, 20, 89–96. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.I. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nussbaumwagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Kirsten, B.; Lewis, L.; John, D. The origin of the naked grains of maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.Y.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. MiR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Stoeckert, C.J., Jr.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.; Rinn, J.; Pachter, L.; et al. Differential gene and transcript expression analysis of RNA-Seq experiments with Hophat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | RefSeq ID | Location | CDS (bp) | Amino Acids Length (aa) | MW (kDa) | pI | GRAVY | Instability Index (II) | EST |
|---|---|---|---|---|---|---|---|---|---|---|
| ZjSBP01 | LOC107425063 | XM_016034996.1 | chr01:21341210-21343540 | 465 | 154 | 17,411.90 | 6.66 | −1.382 | 88.82 | YES |
| ZjSBP02 | LOC107416051 | XM_016024498.1 | chr04:9923829-9929510 | 3180 | 1059 | 117,174.39 | 8.73 | −0.476 | 60.91 | YES |
| ZjSBP03 | LOC107416470 | XM_016024954.1 | chr04:14303210-14305417 | 642 | 213 | 23,753.22 | 9.04 | −1.127 | 60.78 | NO |
| ZjSBP04A | LOC107418038 | XM_016026704.1 | chr05:6738154-6742177 | 1185 | 394 | 42,994.94 | 7.26 | −0.576 | 53.17 | YES |
| ZjSBP04B | LOC107418038 | XM_016026705.1 | chr05:6738154-6742177 | 1185 | 394 | 42,994.94 | 7.26 | −0.576 | 53.17 | YES |
| ZjSBP05 | LOC107419077 | XM_016027806.1 | chr05:21507138-21511275 | 1665 | 554 | 60,692.96 | 8.07 | −0.487 | 51.59 | YES |
| ZjSBP06 | LOC107417566 | XM_016026174.1 | chr05:2335756-2337973 | 1245 | 414 | 45,301.32 | 8.54 | −0.580 | 58.86 | NO |
| ZjSBP07A | LOC107423307 | XM_016032840.1 | chr07:23461384-23466777 | 2487 | 828 | 92,658.80 | 6.20 | −0.427 | 56.10 | YES |
| ZjSBP07B | LOC107423307 | XM_016032841.1 | chr07:23461384-23466777 | 2400 | 799 | 89,422.84 | 6.04 | −0.481 | 55.73 | YES |
| ZjSBP08 | LOC107424290 | XM_016034051.1 | chr08:5519236-5520446 | 519 | 172 | 19,646.28 | 5.20 | −1.309 | 78.87 | YES |
| ZjSBP09A | LOC107426205 | XM_016036321.1 | chr09:3743974-3750726 | 3057 | 1018 | 112,964.47 | 6.22 | −0.482 | 49.87 | YES |
| ZjSBP09B | LOC107426205 | XM_016036322.1 | chr09:3743974-3750726 | 3057 | 1018 | 112,964.47 | 6.22 | −0.482 | 49.87 | YES |
| ZjSBP09C | LOC107426205 | XM_016036323.1 | chr09:3743974-3750726 | 3057 | 1018 | 112,964.47 | 6.22 | −0.482 | 49.87 | YES |
| ZjSBP10 | LOC107429070 | XM_016039720.1 | chr10:13935247-13941596 | 1545 | 514 | 57,356.09 | 7.56 | −0.610 | 56.15 | YES |
| ZjSBP11A | LOC107428311 | XM_016038829.1 | chr10:2775470-2780707 | 1440 | 479 | 52,124.50 | 7.24 | −0.762 | 56.96 | YES |
| ZjSBP11B | LOC107428311 | XM_016038830.1 | chr10:2775470-2780707 | 1440 | 479 | 52,124.50 | 7.24 | −0.762 | 56.96 | YES |
| ZjSBP11C | LOC107428311 | XM_016038831.1 | chr10:2775470-2780707 | 1440 | 479 | 52,124.50 | 7.24 | −0.762 | 56.96 | YES |
| ZjSBP11D | LOC107428311 | XM_016038833.1 | chr10:2775470-2780707 | 1440 | 479 | 52,124.50 | 7.24 | −0.762 | 56.96 | YES |
| ZjSBP12A | LOC107428287 | XM_016038800.1 | chr10:2934047-2937123 | 1065 | 354 | 39,239.78 | 8.54 | −0.723 | 63.10 | YES |
| ZjSBP12B | LOC107428287 | XM_016038801.1 | chr10:2934047-2937123 | 1032 | 343 | 37,911.27 | 8.55 | −0.717 | 62.37 | YES |
| ZjSBP13A | LOC107432336 | XM_016043455.1 | chr12:6956478-6961330 | 1167 | 388 | 41,547.47 | 9.11 | −0.532 | 54.74 | YES |
| ZjSBP13B | LOC107432336 | XM_016043456.1 | chr12:6956478-6961330 | 1146 | 381 | 40,762.53 | 9.11 | −0.542 | 55.78 | YES |
| ZjSBP14 | LOC107433357 | XM_016044643.1 | chr12:19190329-19195805 | 3135 | 1044 | 116,394.53 | 7.84 | −0.419 | 50.17 | YES |
| ZjSBP15 | LOC107406208 | XM_016013312.1 | add_scaffold 414:32884-34897 | 579 | 192 | 21,583.47 | 9.56 | −0.992 | 66.24 | YES |
| ZjSBP16A | LOC107408850 | XM_016016273.1 | add_scaffold2409:1682-4023 | 1167 | 388 | 43,549.82 | 8.29 | −0.743 | 53.37 | YES |
| ZjSBP16B | LOC107408850 | XM_016016274.1 | add_scaffold2409:1682-4023 | 1143 | 380 | 47,233.32 | 8.98 | −0.757 | 52.77 | YES |
| ZjSBP16C | LOC107408850 | XM_016016275.1 | add_scaffold2409:1682-4023 | 1083 | 360 | 40,305.19 | 8.490 | −0.751 | 57.42 | YES |
| ZjSBP16D | LOC107408850 | XM_016016277.1 | add_scaffold2409:1682-4023 | 1083 | 360 | 40,305.19 | 8.490 | −0.751 | 57.42 | YES |
| Motif No. | E-Value | Sites | Width | Annotation of Motif |
|---|---|---|---|---|
| 1 | 1.7 × 10−69 | 16 | 80 | SBP domain |
| 2 | 3.3 × 10−67 | 4 | 68 | unknown |
| 3 | 6.3 × 10−36 | 3 | 78 | Ankyrin repeat |
| 4 | 1.6 × 10−17 | 3 | 33 | unknown |
| 5 | 2.3 × 10−16 | 2 | 78 | unknown |
| 6 | 1.1 × 10−9 | 2 | 70 | unknown |
| 7 | 1.6 × 10−7 | 8 | 11 | unknown |
| 8 | 1.7 × 10−7 | 4 | 19 | unknown |
| 9 | 2.1 × 10−5 | 5 | 19 | unknown |
| 10 | 1.2 × 10−4 | 2 | 53 | unknown |
| Name | E-Value | Best Possible Match |
|---|---|---|
| Motif 1 | 1.7 × 10−69 | PSCQVEGCNADLSSAKDYHRRHKVCELHSKAPKVJVGGLEQRFCQQCSRFHELSEFDEGKRSCRRRLAGHNERRRKPQPE |
| Motif 2 | 3.37 × 10−67 | AQSRTGRIVFKLFGKDPNDFPLVLRAQILDWLSNSPSDIESYIRPGCIILTIYLAMPEAAWEELCENL |
| Motif 3 | 6.37 × 10−36 | AQSRTGRIVFKLFGKDPNDFPLVLRAQILDWLSNSPSDIESYIRPGCIILTIYLAMPEAAWEELCENL |
| Motif 4 | 1.67 × 10−17 | LLYRPAMLSMVAIAAVCVCVALLFKSSPEVVYV |
| Motif 5 | 2.37 × 10−16 | GNIEAKKQALDFIHEMGWLLHRSRAKLRLGHLDPNADPFPFKRFKWLMEFSLEHDWCAVVKKLLGILFEGSVDEGEHP |
| Motif 6 | 1.17 × 10−9 | DPFWRTGWVYIRLQNFIAFIYNGHVIJDTPLPLKSHKNCKILSIKPIAISASEKAQFIVKGFNLARPATR |
| Motif 7 | 1.67 × 10−7 | CALSLLSSQPT |
| Motif 8 | 1.77 × 10−7 | FFPFIVADEEVCSEIRVLE |
| Motif 9 | 2.17 × 10−5 | LEWDLKDWSWDGTLFLAEP |
| Motif 10 | 1.27 × 10−4 | TVVNGNSLNDERGSGYLLISLLRILSNMHSNRSDQNKDQDLLSHLLRSLANFT |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, S.; Zhou, H.; Sheng, S.; Cao, M.; Li, Y.; Pang, X. Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.). Int. J. Mol. Sci. 2017, 18, 1734. https://doi.org/10.3390/ijms18081734
Song S, Zhou H, Sheng S, Cao M, Li Y, Pang X. Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.). International Journal of Molecular Sciences. 2017; 18(8):1734. https://doi.org/10.3390/ijms18081734
Chicago/Turabian StyleSong, Shuang, Heying Zhou, Songbai Sheng, Ming Cao, Yingyue Li, and Xiaoming Pang. 2017. "Genome-Wide Organization and Expression Profiling of the SBP-Box Gene Family in Chinese Jujube (Ziziphus jujuba Mill.)" International Journal of Molecular Sciences 18, no. 8: 1734. https://doi.org/10.3390/ijms18081734