Human Leukocyte Antigen Class I and Class II Polymorphisms and Serum Cytokine Profiles in Cervical Cancer
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Sample Collection
4.3. DNA Extraction and HPV Genotyping
4.4. HLA Class I and II Genotyping
4.5. Serum Cytokine Measurements
4.6. Statistical Analyses
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| CC | Cervical cancer |
| HPV | Human papillomavirus |
| HR-HPV | High-risk human papillomavirus |
| HLA | Human leukocyte antigens |
| Th1 | T-helper 1 |
| Th2 | T-helper 2 |
| Th17 | T-helper 17 |
| IL-6 | Interleukin 6 |
| IL-12 | Interleukin 12 |
| TNF-α | Tumor necrosis factor-α |
| IFN-γ | Interferon-γ |
| IL-4 | Interleukin-4 |
| IL-5 | Interleukin-5 |
| IL-10 | Interleukin-10 |
| IL-13 | Interleukin-13 |
| IL-17 | Interleukin-17 |
| IL-22 | Interleukin-22 |
| NILM | Negative for intraepithelial lesion or malignancy |
| LSIL | Low-grade squamous intraepithelial lesions |
| HSIL | High-grade squamous intraepithelial lesions |
| OR | Odds ratio |
| CI | Confidence interval |
| CTLs | Cytotoxic T cells |
| SD | Standard deviation |
| CIN | Cervical intraepithelial neoplasia |
| ASC | Atypical squamous cells |
| ASC-H | ASC but not possible to exclude HSIL |
| ASC-US | ASC of undetermined significance |
| PCR | Polymerase chain reaction |
| RFLP | Restriction fragment length polymorphism |
| SSO | Sequence-specific oligonucleotide |
| ELISA | Enzyme-linked immunosorbent assay |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Rodríguez, A.C.; Schiffman, M.; Herrero, R.; Wacholder, S.; Hildesheim, A.; Castle, P.E.; Solomon, D.; Burk, R. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J. Natl. Cancer Inst. 2008, 100, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Franco, E.L.; Villa, L.L.; Sobrinho, J.P.; Prado, J.M.; Rousseau, M.C.; Désy, M.; Rohan, T.E. Epidemiology of acquisition and clearance of cervical human papillomavirus infection in women from a high-risk area for cervical cancer. J. Infect. Dis. 1999, 180, 1415–1423. [Google Scholar] [CrossRef] [PubMed]
- Ho, G.Y.; Bierman, R.; Beardsley, L.; Chang, C.J.; Burk, R.D. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 1998, 338, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Tota, J.E.; Chevarie-Davis, M.; Richardson, L.A.; Devries, M.; Franco, E.L. Epidemiology and burden of HPV infection and related diseases: Implications for prevention strategies. Prev. Med. 2011, 53 (Suppl. S1), S12–S21. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.H.; Kim, M.K.; Lee, I.H.; Kim, T.J.; Kwak, S.H.; Song, S.H.; Lee, J.K. Association between serum cytokine profiles and clearance or persistence of high-risk human papillomavirus infection: A prospective study. Int. J. Gynecol. Cancer 2010, 20, 1011–1016. [Google Scholar] [CrossRef] [PubMed]
- Gimenes, F.; Teixeira, J.J.; de Abreu, A.L.; Souza, R.P.; Pereira, M.W.; da Silva, V.R.; Bôer, C.G.; Maria-Engler, S.S.; Bonini, M.G.; Borelli, S.D.; et al. Human leukocyte antigen (HLA)-G and cervical cancer immunoediting: A candidate molecule for therapeutic intervention and prognostic biomarker? Biochim. Biophys. Acta 2014, 1846, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Hildesheim, A.; Wang, S.S. Host and viral genetics and risk of cervical cancer: A review. Virus Res. 2002, 89, 229–240. [Google Scholar] [CrossRef]
- Wu, T.C. Immunology of the human papilloma virus in relation to cancer. Curr. Opin. Immunol. 1994, 6, 746–754. [Google Scholar] [CrossRef]
- Chen, D.; Juko-Pecirep, I.; Hammer, J.; Ivansson, E.; Enroth, S.; Gustavsson, I.; Feuk, L.; Magnusson, P.K.; McKay, J.D.; Wilander, E.; et al. Genome-wide association study of susceptibility loci for cervical cancer. J. Natl. Cancer Inst. 2013, 105, 624–633. [Google Scholar] [CrossRef] [PubMed]
- de Araujo Souza, P.S.; Sichero, L.; Maciag, P.C. HPV variants and HLA polymorphisms: The role of variability on the risk of cervical cancer. Future Oncol. 2009, 5, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Hildesheim, A.; Schiffman, M.; Scott, D.R.; Marti, D.; Kissner, T.; Sherman, M.E.; Glass, A.G.; Manos, M.M.; Lorincz, A.T.; Kurman, R.J.; et al. Human leukocyte antigen class I/II alleles and development of human papillomavirus-related cervical neoplasia: Results from a case-control study conducted in the United States. Cancer Epidemiol. Biomark. Prev. 1998, 7, 1035–1041. [Google Scholar]
- Madeleine, M.M.; Brumback, B.; Cushing-Haugen, K.L.; Schwartz, S.M.; Daling, J.R.; Smith, A.G.; Nelson, J.L.; Porter, P.; Shera, K.A.; McDougall, J.K.; et al. Human leukocyte antigen class II and cervical cancer risk: A population-based study. J. Infect. Dis. 2002, 186, 1565–1574. [Google Scholar] [CrossRef] [PubMed]
- Madeleine, M.M.; Johnson, L.G.; Smith, A.G.; Hansen, J.A.; Nisperos, B.B.; Li, S.; Zhao, L.P.; Daling, J.R.; Schwartz, S.M.; Galloway, D.A. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res. 2008, 68, 3532–3539. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, L.; Hu, Z.; Li, S.; Wang, S.; Liu, J.; Wu, C.; He, L.; Zhou, J.; Li, Z.; et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 2013, 45, 918–922. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Hildesheim, A.; Gao, X.; Schiffman, M.; Herrero, R.; Bratti, M.C.; Sherman, M.E.; Barnes, W.A.; Greenberg, M.D.; McGowan, L.; et al. Comprehensive analysis of human leukocyte antigen class I alleles and cervical neoplasia in 3 epidemiologic studies. J. Infect. Dis. 2002, 186, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Hildesheim, A.; Gao, X.; Schiffman, M.; Herrero, R.; Bratti, M.C.; Sherman, M.E.; Barnes, W.A.; Greenberg, M.D.; McGowan, L.; et al. Human leukocyte antigen class I alleles and cervical neoplasia: No heterozygote advantage. Cancer Epidemiol. Biomark. Prev. 2002, 11, 419–420. [Google Scholar]
- Zhu, J.; Yamane, H.; Paul, W.E. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 2010, 28, 445–489. [Google Scholar] [CrossRef] [PubMed]
- Punt, S.; Fleuren, G.J.; Kritikou, E.; Lubberts, E.; Trimbos, J.B.; Jordanova, E.S.; Gorter, A. Angels and demons: Th17 cells represent a beneficial response, while neutrophil IL-17 is associated with poor prognosis in squamous cervical cancer. Oncoimmunol 2015, 3, e984539. [Google Scholar] [CrossRef] [PubMed]
- Zijlmans, H.J.M.A.A.; Punt, S.; Fleuren, G.J.; Trimbos, J.B.; Kenter, G.G.; Gorter, A. Role of IL-12p40 in cervical carcinoma. Br. J. Cancer 2012, 107, 1956–1962. [Google Scholar]
- Wang, S.S.; Wheeler, C.M.; Hildesheim, A.; Schiffman, M.; Herrero, R.; Bratti, M.C.; Sherman, M.E.; Alfaro, M.; Hutchinson, M.L.; Morales, J.; et al. Human leukocyte antigen class I and II alleles and risk of cervical neoplasia: Results from a population-based study in Costa Rica. J. Infect. Dis. 2001, 184, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Sato, A. The HLA system. First of two parts. N. Engl. J. Med. 2000, 343, 702–709. [Google Scholar] [PubMed]
- Mota, F.; Rayment, N.; Chong, S.; Singer, A.; Chain, B. The antigen-presenting environment in normal and human papillomavirus (HPV)-related premalignant cervical epithelium. Clin. Exp. Immunol. 1999, 116, 33–40. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. Papillomaviruses and cancer: From basic studies to clinical application. Nat. Rev. Cancer 2002, 2, 342–350. [Google Scholar] [CrossRef] [PubMed]
- Bailey, S.R.; Nelson, M.H.; Himes, R.A.; Li, Z.; Mehrotra, S.; Paulos, C.M. Th17 cells in cancer: The ultimate identity crisis. Front. Immunol. 2014, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Paradkar, P.H.; Joshi, J.V.; Mertia, P.N.; Agashe, S.V.; Vaidya, R.A. Role of cytokines in genesis, progression and prognosis of cervical cancer. Asian Pac. J. Cancer Prev. 2014, 15, 3851–3864. [Google Scholar] [CrossRef] [PubMed]
- de Abreu, A.L.; Malaguti, N.; Souza, R.P.; Uchimura, N.S.; Ferreira, É.; Pereira, M.W.; Carvalho, M.D.; Pelloso, S.M.; Bonini, M.G.; Gimenes, F.; et al. Association of human papillomavirus, Neisseria gonorrhoeae and Chlamydia trachomatis co-infections on the risk of high-grade squamous intraepithelial cervical lesion. Am. J. Cancer Res. 2016, 6, 1371–1383. [Google Scholar] [PubMed]
- Mege, J.L.; Meghari, S.; Honstettre, A.; Capo, C.; Raoult, D. The two faces of interleukin 10 in human infectious diseases. Lancet Infect. Dis. 2006, 6, 557–569. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.H.; Li, Y.H.; Li, O. The paradox of IL-10-mediated modulation in cervical cancer. Biomed. Rep. 2013, 1, 347–351. [Google Scholar] [PubMed]
- Santin, A.D.; Hermonat, P.L.; Ravaggi, A.; Bellone, S.; Pecorelli, S.; Roman, J.J.; Parham, G.P.; Cannon, M.J. Interleukin-10 increases Th1 cytokine production and cytotoxic potential in human papillomavirus-specific CD8(+) cytotoxic T lymphocytes. J. Virol. 2000, 74, 4729–4737. [Google Scholar] [CrossRef] [PubMed]
- Kohno, T.; Mizukami, H.; Suzuki, M.; Saga, Y.; Takei, Y.; Shimpo, M.; Matsushita, T.; Okada, T.; Hanazono, Y.; Kume, A.; et al. Interleukin-10-mediated inhibition of angiogenesis and tumor growth in mice bearing VEGF-producing ovarian cancer. Cancer Res. 2003, 63, 5091–5094. [Google Scholar] [PubMed]
- Castrilli, G.; Tatone, D.; Diodoro, M.G.; Rosini, S.; Piantelli, M.; Musiani, P. Interleukin 1alpha and interleukin 6 promote the in vitro growth of both normal and neoplastic human cervical epithelial cells. Br. J. Cancer 1997, 75, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.H.; Kuo, M.L.; Chen, C.A.; Chou, C.H.; Lai, K.B.; Lee, C.N.; Hsieh, C.Y. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene 2003, 22, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Davidson, E.J.; Davidson, J.A.; Sterling, J.C.; Baldwin, P.J.; Kitchener, H.C.; Stern, P.L. Association between human leukocyte antigen polymorphism and human papillomavirus 16-positive vulval intraepithelial neoplasia in British women. Cancer Res. 2003, 63, 400–403. [Google Scholar] [PubMed]
- Qiu, X.; Zhang, F.; Chen, D.; Azad, A.K.; Zhang, L.; Yuan, Y.; Jiang, Z.; Liu, W.; Tan, Y.; Tao, N. HLA-B*07 is a high risk allele for familial cervical cancer. Asian Pac. J. Cancer Prev. 2011, 12, 2597–2600. [Google Scholar] [PubMed]
- Chuang, L.C.; Hu, C.Y.; Chen, H.C.; Lin, P.J.; Lee, B.; Lin, C.Y.; Pan, M.H.; You, S.L.; Hsieh, C.Y.; Chen, C.J.; et al. Associations of human leukocyte antigen class II genotypes with human papillomavirus 18 infection and cervical intraepithelial neoplasia risk. Cancer 2012, 118, 223–231. [Google Scholar] [CrossRef] [PubMed]
- De Araujo Souza, P.S.; Villa, L.L. Genetic susceptibility to infection with human papillomavirus and development of cervical cancer in women in Brazil. Mutat. Res. 2003, 544, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Ivansson, E.L.; Magnusson, J.J.; Magnusson, P.K.; Erlich, H.A.; Gyllensten, U.B. MHC loci affecting cervical cancer risk: Distinguishing the effects of HLA-DQB1 and non-hla genes TNF, LTA, TAP1 and TAP2. Genes Immun. 2008, 9, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Xu, A.; Xie, Y.; Awonuga, A.O.; Lin, Z. Some but not all of HLA-II alleles are associated with cervical cancer in Chinese women. Cancer Genet. Cytogenet. 2008, 187, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, L.; Tian, C.; Yang, L.; Wang, Z. Genetic variants and risk of cervical cancer: Epidemiological evidence, meta-analysis and research review. BJOG 2014, 121, 664–674. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.P.; Cheung, T.H.; Tam, A.O.; Cheung, J.L.; Yim, S.F.; Lo, K.W.; Siu, N.S.; Zhou, D.X.; Chan, P.K. Risk association between human leukocyte antigen-A allele and high-risk human papillomavirus infection for cervical neoplasia in Chinese women. J. Infect. Dis. 2005, 192, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Hosono, S.; Kawase, T.; Matsuo, K.; Watanabe, M.; Kajiyama, H.; Hirose, K.; Suzuki, T.; Kidokoro, K.; Ito, H.; Nakanishi, T.; et al. HLA-A alleles and the risk of cervical squamous cell carcinoma in Japanese women. J. Epidemiol. 2010, 20, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.K.S.; Cheung, J.L.K.; Cheung, T.-H.; Lin, C.K.; Siu, S.-S.N.; Yu, M.M.Y.; Tang, J.W.; Lo, K.W.K.; Yim, S.-F.; Wong, Y.F.; et al. HLA-DQB1 polymorphisms and risk of cervical cancer: A case-control study in a southern Chinese population. Gynecol. Oncol. 2007, 105, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, Y.; Li, L.; Cao, Y.; Liu, Z.; Liu, B.; Du, Z.; Zhang, Y.; Chen, S.; Lin, Z.; et al. Polymorphic amino acids at codons 9 and 37 of HLA-DQB1 alleles may confer susceptibility to cervical cancer among Chinese women. Int. J. Cancer 2006, 118, 3006–3011. [Google Scholar] [CrossRef] [PubMed]
- Beskow, A.H.; Gyllensten, U.B. Host genetic control of HPV 16 titer in carcinoma in situ of the cervix uteri. Int. J. Cancer 2002, 101, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, J.; Terry, G.; Ho, L.; Monaghan, J.; Lopes, A.; Clarkson, P.; Duncan, I. Association between high-risk HPV types, HLA DRB1* and DQB1* alleles and cervical cancer in British women. Br. J. Cancer 2000, 82, 1348–1352. [Google Scholar] [PubMed]
- De Araujo Souza, P.S.; Maciag, P.C.; Ribeiro, K.B.; Petzl-Erler, M.L.; Franco, E.L.; Villa, L.L. Interaction between polymorphisms of the human leukocyte antigen and HPV-16 variants on the risk of invasive cervical cancer. BMC Cancer 2008, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Maciag, P.C.; Schlecht, N.F.; Souza, P.S.; Franco, E.L.; Villa, L.L.; Petzl-Erler, M.L. Major histocompatibility complex class II polymorphisms and risk of cervical cancer and human papillomavirus infection in Brazilian women. Cancer Epidemiol. Biomark. Prev. 2000, 9, 1183–1191. [Google Scholar]
- Maciag, P.C.; Schlecht, N.F.; Souza, P.S.; Rohan, T.E.; Franco, E.L.; Villa, L.L. Polymorphisms of the human leukocyte antigen DRB1 and DQB1 genes and the natural history of human papillomavirus infection. J. Infect. Dis. 2002, 186, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Koutsky, L.A.; Critchlow, C.W.; Apple, R.J.; Hawes, S.E.; Hughes, J.P.; Touré, P.; Dembele, A.; Kiviat, N.B. HLA class II DR-DQ and increased risk of cervical cancer among Senegalese women. Cancer Epidemiol. Biomark. Prev. 2001, 10, 1037–1045. [Google Scholar]
- Marangon, A.V.; Guelsin, G.A.; Visentainer, J.E.; Borelli, S.D.; Watanabe, M.A.; Consolaro, M.E.; Caleffi-Ferracioli, K.R.; Rudnick, C.C.; Sell, A.M. The association of the immune response genes to human papillomavirus-related cervical disease in a Brazilian population. BioMed Res. Int. 2013, 2013, 146079. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Hildesheim, A. Viral and host factors in human papillomavirus persistence and progression. J. Natl. Cancer Inst. Monogr. 2003, 35–40. [Google Scholar] [CrossRef]
- Bruni, L.; Diaz, M.; Castellsagué, X.; Ferrer, E.; Bosch, F.X.; de Sanjosé, S. Cervical human papillomavirus prevalence in 5 continents: Meta-analysis of 1 million women with normal cytological findings. J. Infect. Dis. 2010, 202, 1789–1799. [Google Scholar]
- Human papillomavirus vaccines: WHO position paper, May 2017. Wkly. Epidemiol. Rec. 2017, 92, 241–268.
- Serrano, B.; de Sanjosé, S.; Tous, S.; Quiros, B.; Muñoz, N.; Bosch, X.; Alemany, L. Human papillomavirus genotype attribution for HPVs 6, 11, 16, 18, 31, 33, 45, 52 and 58 in female anogenital lesions. Eur. J. Cancer 2015, 51, 1732–1741. [Google Scholar] [CrossRef] [PubMed]
- Probst, C.M.; Bompeixe, E.P.; Pereira, N.F.; de O’Dalalio, M.M.; Visentainer, J.E.; Tsuneto, L.T.; Petzl-Erler, M.L. HLA polymorphism and evaluation of European, African, and Amerindian contribution to the white and mulatto populations from Paraná, Brazil. Hum. Biol. 2000, 72, 597–617. [Google Scholar] [PubMed]
- Sobin, L.H. The international histological classification of tumours. Bull. World Health Organ. 1981, 59, 813–819. [Google Scholar] [PubMed]
- Katz, L.M.C. Review of the Brazilian guidelines for cervical cancer screening, 2016. J. Bras. Patol. Med. Lab. 2016, 52, 210. [Google Scholar] [CrossRef]
- Chen, L.; Watanabe, K.; Haruyama, T.; Kobayashi, N. Simple and rapid human papillomavirus genotyping method by restriction fragment length polymorphism analysis with two restriction enzymes. J. Med. Virol. 2013, 85, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Santiago, E.; Camacho, L.; Junquera, M.L.; Vázquez, F. Full HPV typing by a single restriction enzyme. J. Clin. Virol. 2006, 37, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, K.M.; Dean, A.; Soe, M.M. Openepi: A web-based epidemiologic and statistical calculator for public health. Public Health Rep. 2009, 124, 471–474. [Google Scholar] [CrossRef] [PubMed]
- Ayres, M.; Ayres Júnior, M.; Ayres, D.L.; Santos, A.S.D. Bioestat 5.0: Aplicações Estatísticas nas Áreas das Ciências Biológicas e Médicas, 5th ed.; MCTI/IDSM/CNPq: Belém, Brazil, 2007; pp. 1–364. ISBN 858-5-92-410-1. [Google Scholar]

| Cytology | Women | HPV Positivity | ||
|---|---|---|---|---|
| HPV-DNA | HR-HPV | HPV-16 | ||
| n (%) | n (%) | n (%) | n (%) | |
| NILM | 48 (38.7) | 3 (6.3) | 2 (66.7) | 0 (0.0) |
| LSIL | 27 (21.8) | 26 (96.3) | 12 (46.2) | 06 (23.1) |
| HSIL | 42 (33.9) | 42 (100.0) | 41 (97.6) | 19 (45.2) |
| CC | 07 (5.6) | 07 (100.0) | 07 (100.0) | 04 (57.1) |
| Total | 124 (100.0) | 78 (62.9) | 62 (79.5) | 29 (37.2) |
| HLA-A | n (f%) | B*40 | 17 (6.9) | HLA-DRB1 | n (f%) |
|---|---|---|---|---|---|
| A*01 | 28 (11.3) | B*41 | 1 (0.4) | DRB1*01 | 31 (12.5) |
| A*02 | 69 (27.8) | B*42 | 1 (0.4) | DRB1*03 | 26 (10.5) |
| A*03 | 21 (8.5) | B*44 | 29 (11.7) | DRB1*04 | 24 (9.7) |
| A*11 | 18 (7.3) | B*45 | 3 (1.2) | DRB1*07 | 33 (13.3) |
| A*23 | 9 (3.6) | B*48 | 4 (1.6) | DRB1*08 | 27 (10.9) |
| A*24 | 28 (11.3) | B*49 | 4 (1.6) | DRB1*09 | 6 (2.4) |
| A*25 | 3 (1.2) | B*50 | 4 (1.6) | DRB1*10 | 3 (1.2) |
| A*26 | 9 (3.6) | B*51 | 16 (6.5) | DRB1*11 | 21 (8.5) |
| A*29 | 14 (5.7) | B*52 | 7 (2.8) | DRB1*12 | 3 (1.2) |
| A*30 | 3 (1.2) | B*55 | 2 (0.8) | DRB1*13 | 24 (9.7) |
| A*31 | 24 (9.7) | B*56 | 4 (1.6) | DRB1*14 | 10 (4.0) |
| A*32 | 8 (3.2) | B*57 | 6 (2.4) | DRB1*15 | 28 (11.3) |
| A*33 | 2 (0.8) | B*58 | 5 (2.0) | DRB1*16 | 12 (4.8) |
| A*66 | 3 (1.2) | HLA-C | n (f%) | HLA-DQA1 | n (f%) |
| A*68 | 7 (2.8) | C*01 | 10 (4.0) | DQA1*01 | 89 (35.9) |
| A*74 | 2 (0.8) | C*02 | 13 (5.2) | DQA1*02 | 34 (13.7) |
| HLA-B | n (f%) | C*03 | 40 (16.1) | DQA1*03 | 31 (12.5) |
| B*07 | 21 (8.5) | C*04 | 39 (15.7) | DQA1*04 | 26 (10.5) |
| B*08 | 22 (8.9) | C*05 | 11 (4.4) | DQA1*05 | 67 (27.0) |
| B*13 | 4 (1.6) | C*06 | 17 (6.9) | DQA1*06 | 1 (0.4) |
| B*14 | 9 (3.6) | C*07 | 68 (27.4) | HLA-DQB1 | n (f%) |
| B*15 | 28 (11.3) | C*08 | 9 (3.6) | DQB1*01 | 2 (0.8) |
| B*18 | 10 (4.0) | C*12 | 13 (5.2) | DQB1*02 | 55(22.2) |
| B*27 | 6 (2.4) | C*14 | 1 (0.4) | DQB1*03 | 73 (29.4) |
| B*35 | 28 (11.3) | C*15 | 10 (4.0) | DQB1*04 | 21 (8.5) |
| B*37 | 2 (0.8) | C*16 | 12 (4.8) | DQB1*05 | 49 (19.8) |
| B*38 | 5 (2.0) | C*17 | 3 (1.2) | DQB1*06 | 48 (19.4) |
| B*39 | 10 (4.0) | C*18 | 2 (0.8) |
| HLA Allelic Group | n (ƒ%) | n (ƒ%) | p-Value | OR CI (95%) |
|---|---|---|---|---|
| HPV-negative (n = 46) | HPV-positive (n = 78) | |||
| B*07 | 3 (3.26) | 18 (11.54) | 0.0316 1 | 3.87 (1.11–13.52) |
| HR-HPV-negative (n = 63) | HR-HPV-positive (n = 61) | |||
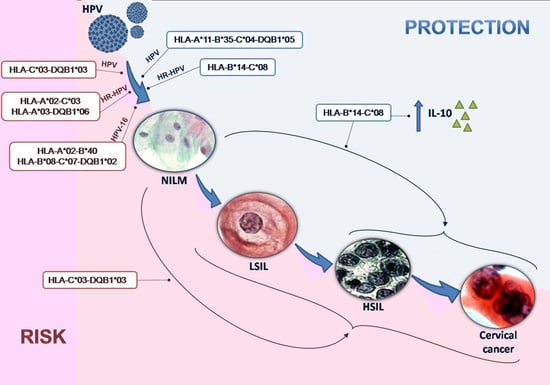
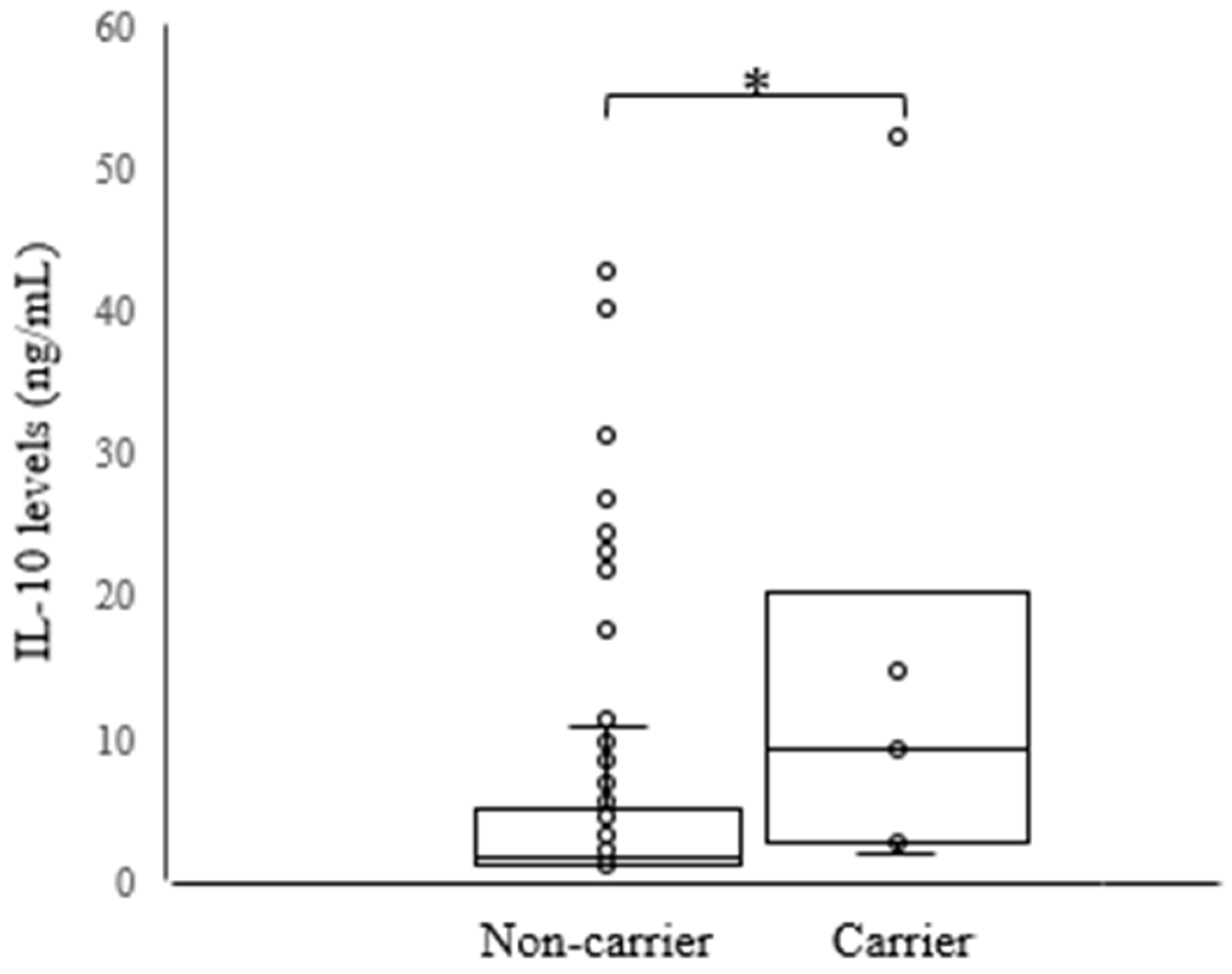
| B*14 | 8 (6.35) | 1 (0.82) | 0.0359 1 | 0.12 (0.15–0.99) |
| C*08 | 8 (6.35) | 1 (0.82) | 0.0359 1 | 0.12 (0.15–0.99) |
| NILM (n = 48) | HSIL/CC (n = 49) | |||
| B*14 | 6 (6.25) | 0 (0.0) | 0.0271 1 | - |
| C*08 | 6 (6.25) | 0 (0.0) | 0.0271 1 | - |
| DQB1*03 | 20 (20.83) | 35 (35.71) | 0.0324 2 | 2.11 (1.11–4.02) |
| NILM (n = 48) | LSIL/HSIL/CC (n = 76) | |||
| DQB1*03 | 20 (20.83) | 53 (34.87) | 0.0265 2 | 2.03 (1.12–3.69) |
| HLA Haplotypes | HPV | HR-HPV | HPV-16 | NILM (n = 48) | LSIL (n = 27) | HSIL/CC (n = 49) | |||
|---|---|---|---|---|---|---|---|---|---|
| Yes (n = 78) | No (n = 46) | Yes (n = 61) | No (n = 63) | Yes (n = 29) | No (n = 95) | ||||
| n (f%) * | n (f%) * | n (f%) * | n (f%) * | n (f%) * | n (f%) * | n (f%) * | n (f%) † | n (f%) † | |
| A*11-B*35-C*04-DRB1*01-DQA1*01-DQB1*05 | 0 (0.0) a | 6 (6.2) a | 0 (0.0) | 6 (4.8) | 0 (0.0) | 6 (3.0) | 6 (5.9) b | 0 (0.0) b | 0 (0.0) b |
| A*11-B*35-C*04-DQB1*05 | 0 (0.0) a | 6 (6.2) a | 0 (0.0) | 6 (4.8) | 0 (0.0) | 6 (3.0) | 6 (5.9) b | 0 (0.0) b | 0 (0.0) b |
| A*11-B*35-C*04 | 0 (0.0) a | 6 (6.2) a | 0 (0.0) | 7 (5.4) | 0 (0.0) | 7 (3.5) | 6 (5.9) b | 0 (0.0) b | 0 (0.0) b |
| A*11-B*35-DQB1*05 | 0 (0.0) a | 6 (6.2) a | 0 (0.0) | 6 (4.8) | 0 (0.0) | 6 (3.0) | 6 (6.3) b | 0 (0.0) b | 0 (0.0) b |
| A*11-C *04-DQB1*05 | 0 (0.0) a | 6 (6.2) a | 0 (0.0) | 6 (4.8) | 0 (0.0) | 6 (3.0) | 6 (6.3) b | 0 (0.0) b | 0 (0.0) b |
| A*01-B*08-C*07-DQB1*02 | 0 (0.0) | 4 (4.3) | 0 (0.0) c | 8 (6.3) c | 0 (0.0) d | 10 (5.1) d | 4 (4.2) | 0 (0.0) | 1 (1.0) |
| B 08-C*07-DQB1*02 | 10 (6.4) | 4 (4.3) | 0 (0.0) c | 10 (7.9) c | 0 (0.0) | 13 (6.8) | 4 (4.2) | 5 (9.2) | 5 (5.1) |
| B*15-C*03-DQB1*03 | 12 (7.8) a | 0 (0.0) a | 9 (7.4) | 5 (4.0) | 4 (6.9) | 10 (5.1) | 0 (0.0) b,e | 4 (7.4) b | 8 (8.1) b,e |
| C*03-DQB1*03 | 16 (10.0) a | 0 (0.0) a | 12 (9.9) | 3 (2.7) | 6 (9.9) | 10 (5.4) | 0 (0.0) b,e | 3 (5.6) b | 12 (12.7) b,e |
| A*02-B*40 | 9 (5.8) | 0 (0.0) | 9 (7.3) c | 0 (0.0) c | 5 (8.6) | 4 (2.1) | 0 (0.0) | 0 (0.0) | 8 (7.9) |
| A*02-C*03 | 11 (7.1) | 2 (2.1) | 11 (9.0) | 3 (2.3) | 9 (15.5) d,† | 8 (4.4) d,‡ | 2 (1.9) | 2 (3.7) | 11 (11.2) |
| A*03-DQB1*06 | 9 (5.7) | 2 (2.2) | 9 (7.8) c | 0 (0.0) c | 7 (11.6) d,‡ | 3 (1.7) d,‡ | 2 (2.1) | 4 (7.4) | 5 (4.7) |
| B*14-C*08 | 3 (1.9) | 6 (6.5) | 1 (0.8) c | 8 (6.4) c | 0 (0.0) | 9 (4.7) | 6 (6.3) e | 3 (5.6) | 0 (0.0) e |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahls, L.; Yamakawa, R.; Zanão, K.; Alfieri, D.; Flauzino, T.; Delongui, F.; De Abreu, A.; Souza, R.; Gimenes, F.; Reiche, E.; et al. Human Leukocyte Antigen Class I and Class II Polymorphisms and Serum Cytokine Profiles in Cervical Cancer. Int. J. Mol. Sci. 2017, 18, 1478. https://doi.org/10.3390/ijms18091478
Bahls L, Yamakawa R, Zanão K, Alfieri D, Flauzino T, Delongui F, De Abreu A, Souza R, Gimenes F, Reiche E, et al. Human Leukocyte Antigen Class I and Class II Polymorphisms and Serum Cytokine Profiles in Cervical Cancer. International Journal of Molecular Sciences. 2017; 18(9):1478. https://doi.org/10.3390/ijms18091478
Chicago/Turabian StyleBahls, Larissa, Roger Yamakawa, Karina Zanão, Daniela Alfieri, Tamires Flauzino, Francieli Delongui, André De Abreu, Raquel Souza, Fabrícia Gimenes, Edna Reiche, and et al. 2017. "Human Leukocyte Antigen Class I and Class II Polymorphisms and Serum Cytokine Profiles in Cervical Cancer" International Journal of Molecular Sciences 18, no. 9: 1478. https://doi.org/10.3390/ijms18091478