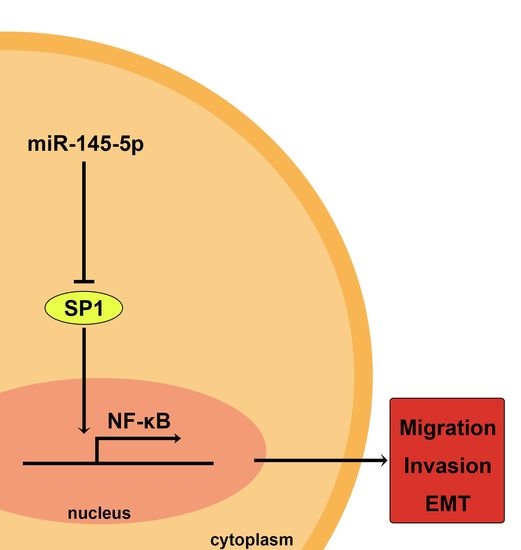
miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Results
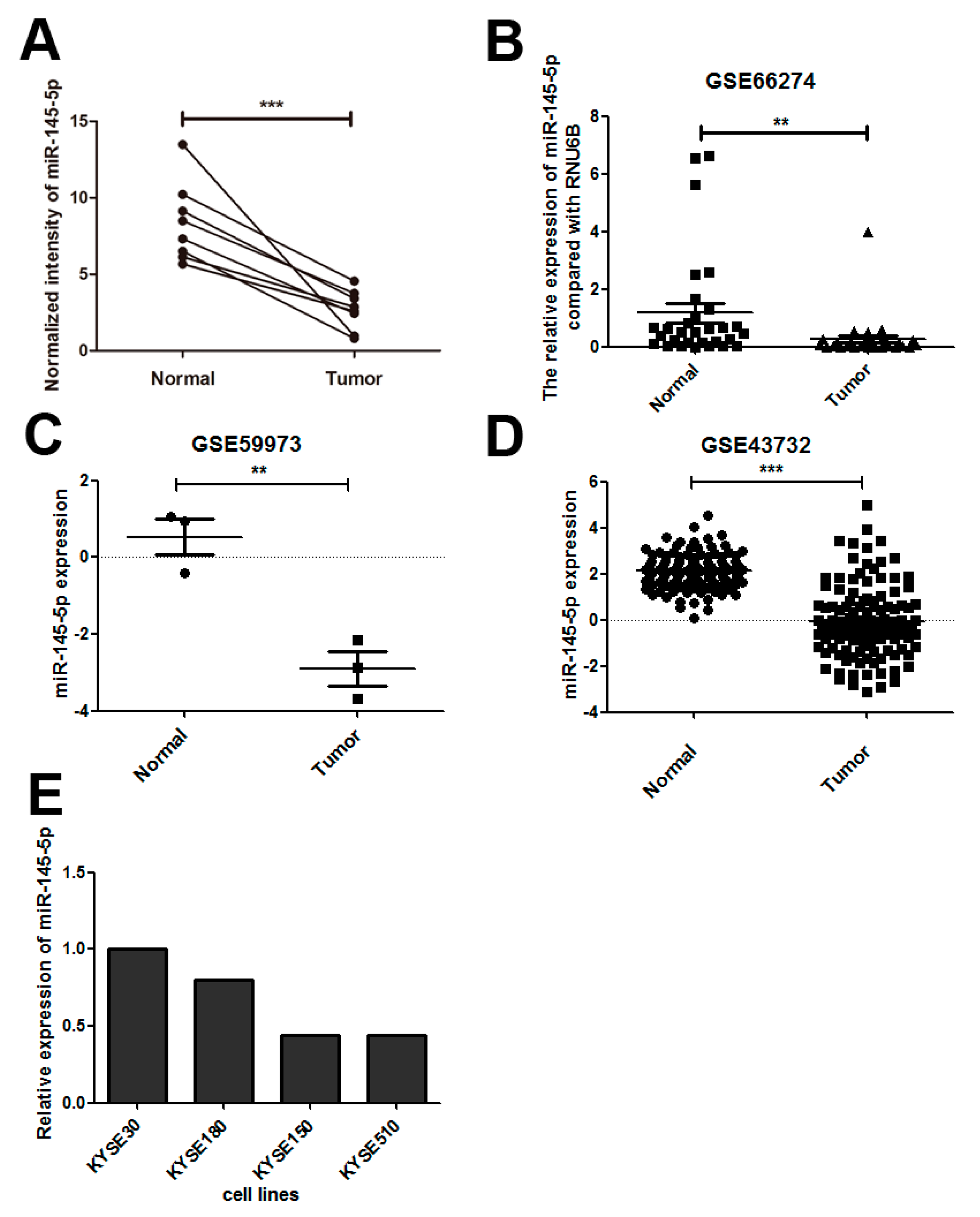
2.1. miR-145-5p Is Down-Regulated in Esophageal Squamous Cell Carcinoma (ESCC) Tissues
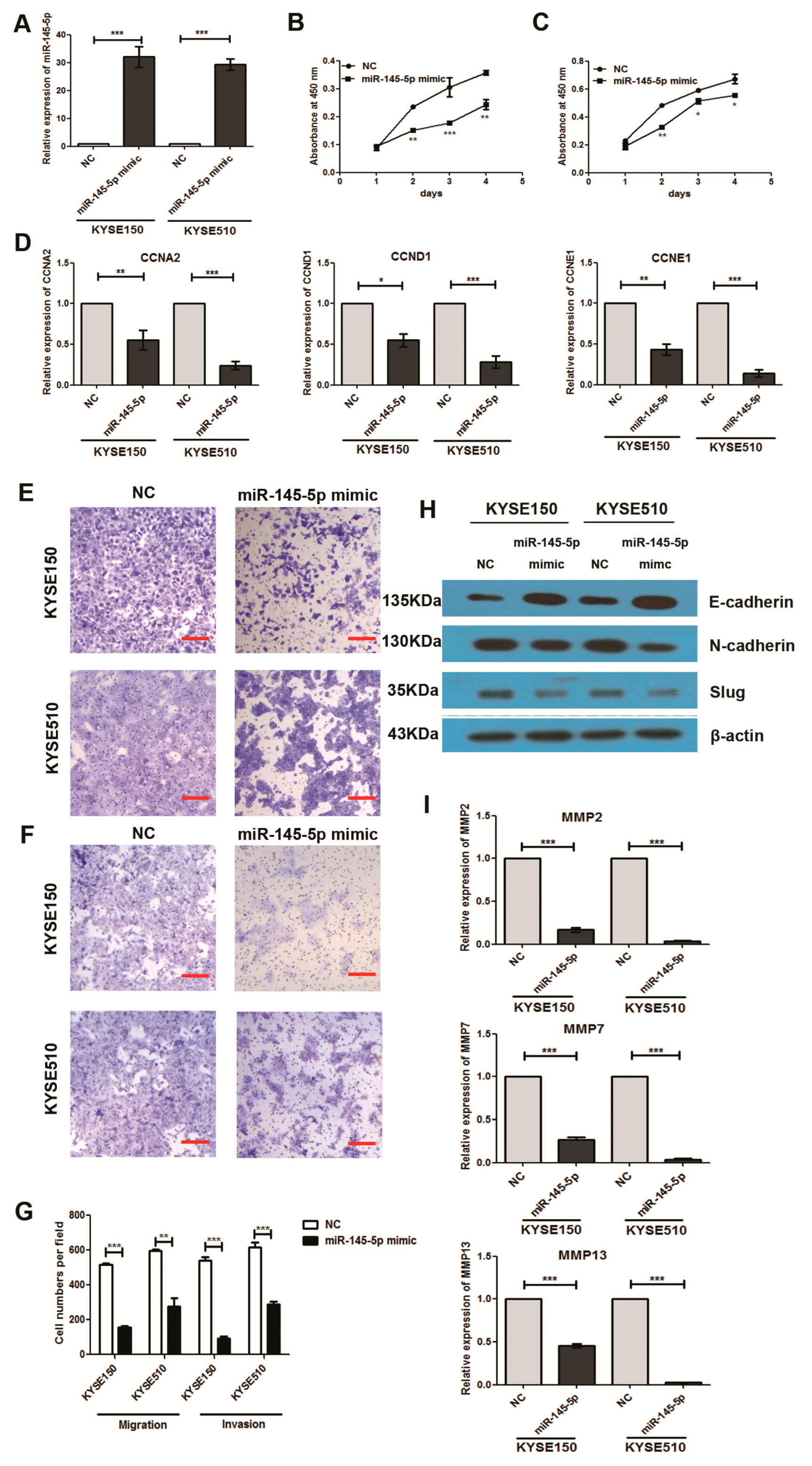
2.2. Overexpression of miR-145-5p Inhibits Cell Proliferation, Migration, Invasion and EMT of ESCC Cells
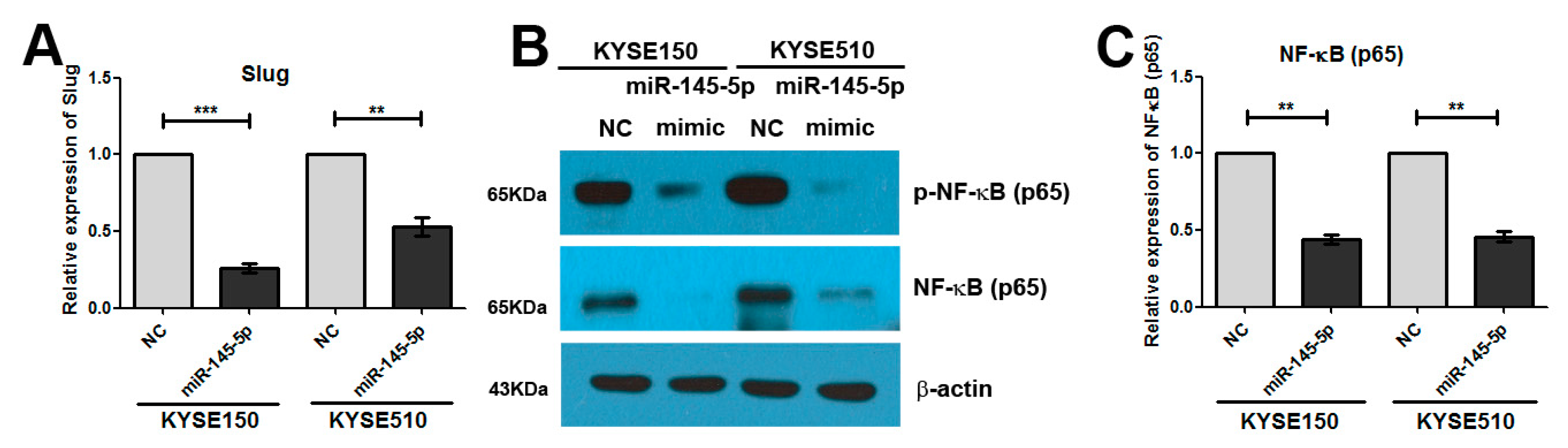
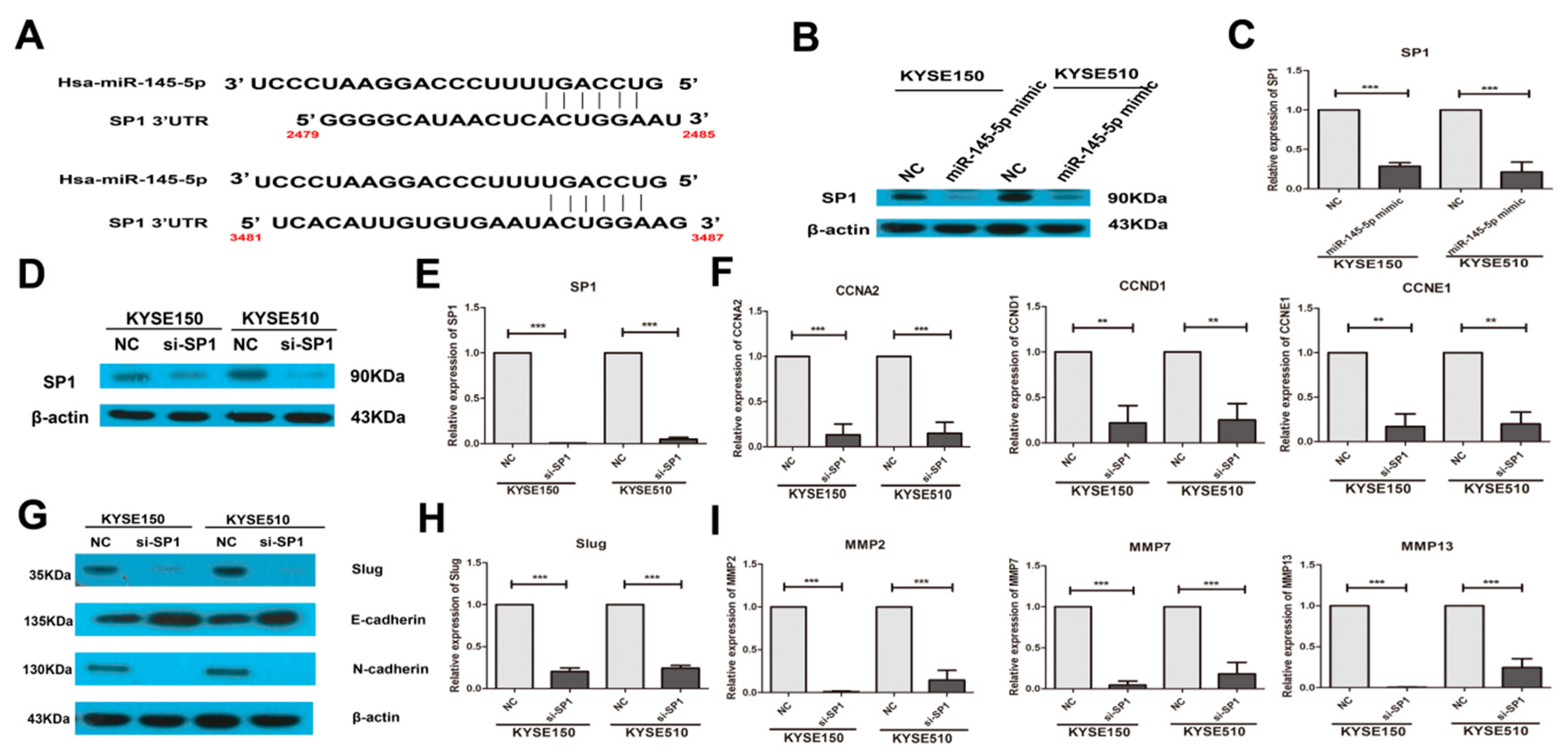
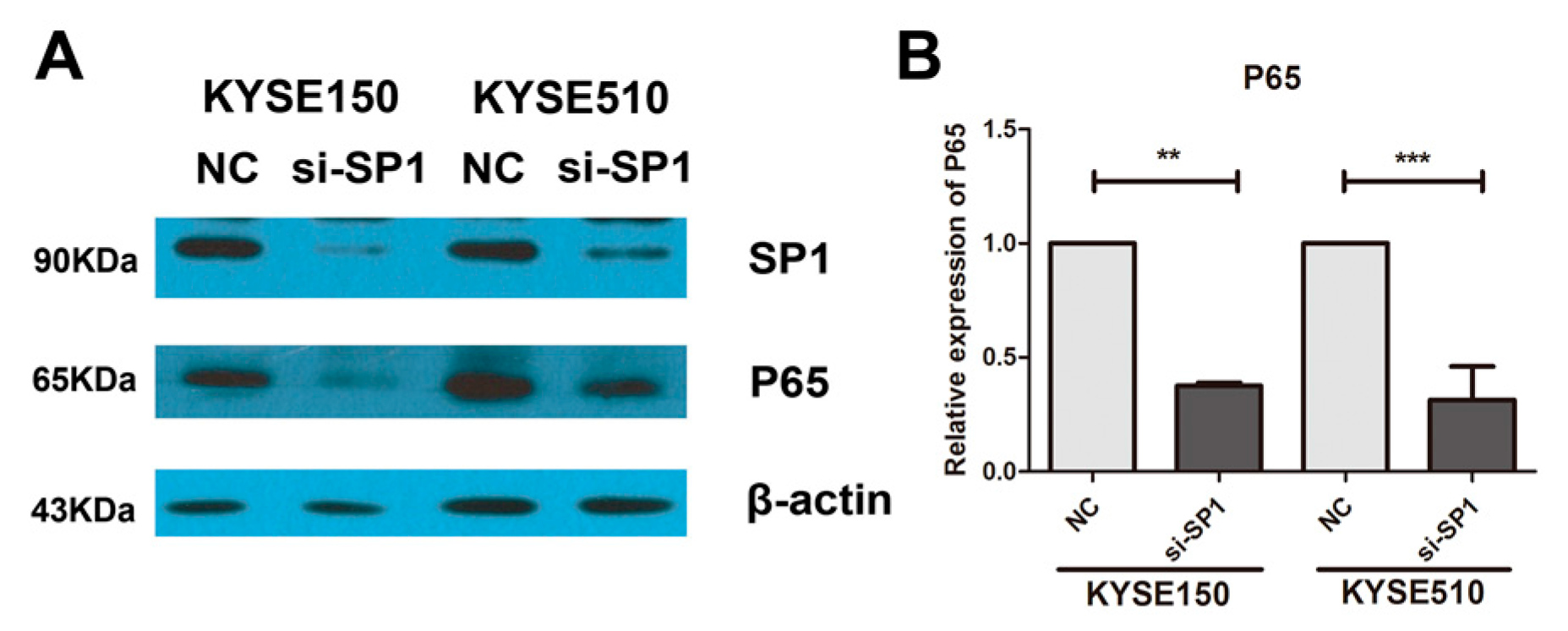
2.3. Overexpression of miR-145-5p Inhibits the Transcription of Slug via the Sp1/NF-κB Signaling Pathway
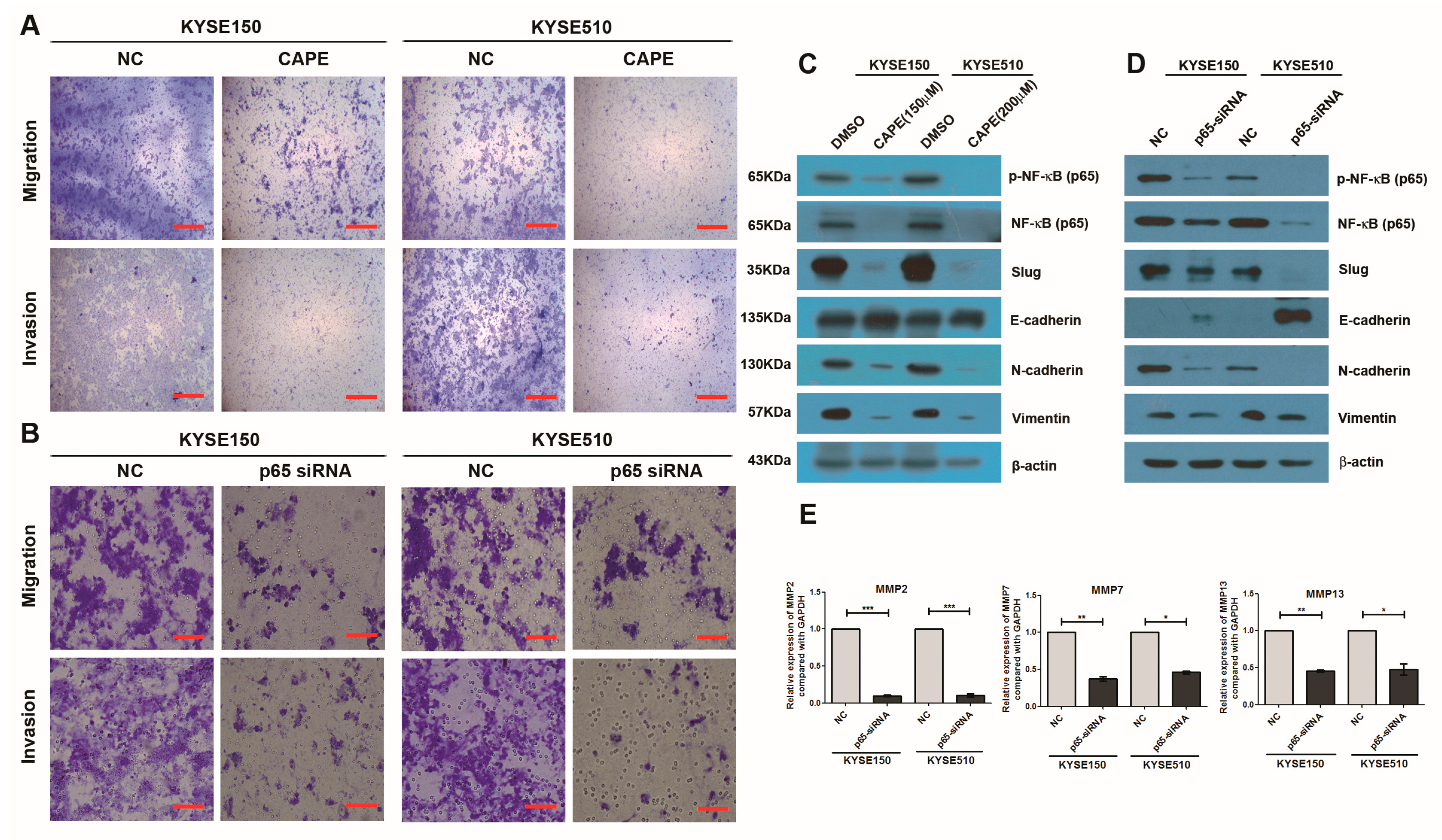
2.4. Inhibition of NF-κB Signaling Pathway or Knockdown of NF-κB (p65) Phenocopied the Effects of miR-145-5p on the Migration, Invasion and EMT of ESCC Cells
3. Discussion
4. Materials and Methods
4.1. Sample Information and miRNA Expression Microarray Detection
4.2. Gene Expression Omnibus (GEO) Datasets
4.3. Cell Culture
4.4. Cell Transfection
4.5. Cell Proliferation Assay
4.6. Cell Migration and Invasion Assay
4.7. Total RNA Extraction and Real-Time PCR Assay
4.8. Western Blotting Assay
4.9. Statistical Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.B.; Li, S.; Li, T.T.; Peng, H.; Jin, T.T.; Zhang, S.M.; Liu, C.X.; Yang, L.; Shen, Y.Y.; Li, S.; et al. Targeting oncogenic PLCE1 by miR-145 impairs tumor proliferation and metastasis of esophageal squamous cell carcinoma. Oncotarget 2016, 7, 1777–1795. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, M.; Khalili, M.; Vasei, M.; Nouraei, N.; Mansour Samaei, N.; Khavanin, A.; Khajehei, M.; Mowla, S.J. Evaluating the miR-302b and miR-145 expression in formalin-fixed paraffin-embedded samples of esophageal squamous cell carcinoma. Arch. Iran. Med. 2015, 18, 173–178. [Google Scholar] [PubMed]
- Wang, F.; Xia, J.; Wang, N.; Zong, H. miR-145 inhibits proliferation and invasion of esophageal squamous cell carcinoma in part by targeting c-Myc. Onkologie 2013, 36, 754–758. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Meng, L.; Wang, L.; Dong, W.; Shen, H.; Wang, G.; Liu, Q.; Du, J. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene 2013, 517, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhang, H.Y.; Zhong, B.L.; Wang, X.J.; Zhang, B.; Chen, H. MicroRNA-145 Inhibits Cell Migration and Invasion and Regulates Epithelial-Mesenchymal Transition (EMT) by Targeting Connective Tissue Growth Factor (CTGF) in Esophageal Squamous Cell Carcinoma. Med. Sci. Monit. 2016, 22, 3925–3934. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Du, A.L.; Wang, L.; Wang, X.Y.; Gao, J.H.; Wang, T.Y. Episomal Lentiviral Vector-Mediated miR-145 Overexpression Inhibits Proliferation and Induces Apoptosis of Human Esophageal Carcinomas Cells. Recent Pat. Anticancer Drug Discov. 2016, 11, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Baba, Y.; Ishimoto, T.; Kosumi, K.; Tokunaga, R.; Izumi, D.; Ohuchi, M.; Nakamura, K.; Kiyozumi, Y.; Kurashige, J.; et al. Suppressor microRNA-145 Is Epigenetically Regulated by Promoter Hypermethylation in Esophageal Squamous Cell Carcinoma. Anticancer Res. 2015, 35, 4617–4624. [Google Scholar] [PubMed]
- Li, J.; Zhu, S.C.; Li, S.G.; Zhao, Y.; Xu, J.R.; Song, C.Y. TKTL1 promotes cell proliferation and metastasis in esophageal squamous cell carcinoma. Biomed. Pharmacother 2015, 74, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, X.; Li, C.; He, C.; Ren, B.; Deng, Q.; Gao, W.; Wang, B. Aurora-A modulates MMP-2 expression via AKT/NF-κB pathway in esophageal squamous cell carcinoma cells. Acta Biochim Biophys Sin. 2016, 48, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.D.; Li, J.Y.; Li, M.; Gu, J.; Shi, X.T.; Ke, Y.; Chen, K.N. Matrix metalloproteinases expression correlates with survival in patients with esophageal squamous cell carcinoma. Am. J. Gastroenterol. 2005, 100, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Khan, P.; Manna, A.; Saha, S.; Mohanty, S.; Mukherjee, S.; Mazumdar, M.; Guha, D.; Das, T. Aspirin inhibits epithelial-to-mesenchymal transition and migration of oncogenic K-ras-expressing non-small cell lung carcinoma cells by down-regulating E-cadherin repressor Slug. BMC Cancer 2016, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Yurochko, A.D.; Mayo, M.W.; Poma, E.E.; Baldwin, A.S., Jr.; Huang, E.S. Induction of the transcription factor Sp1 during human cytomegalovirus infection mediates upregulation of the p65 and p105/p50 NF-κB promoters. J. Virol. 1997, 71, 4638–4648. [Google Scholar] [PubMed]
- Gu, L.; Findley, H.W.; Zhou, M. MDM2 induces NF-kappaB/p65 expression transcriptionally through Sp1-binding sites: A novel, p53-independent role of MDM2 in doxorubicin resistance in acute lymphoblastic leukemia. Blood 2002, 99, 3367–3375. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Zhou, X.; Wang, J.; Du, Y.; Xu, J.; Huang, Z.; Zhu, W.; Shu, Y.; Liu, P. miR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014, 588, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Li, Y.; Xie, C.; Yin, X.; Liu, Y.; Cao, Y.; Fang, Y.; Lin, X.; Xu, Y.; Xu, W.; et al. miR-145 sensitizes ovarian cancer cells to paclitaxel by targeting Sp1 and Cdk6. Int. J. Cancer 2014, 135, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wen-Ming, C.; Jun-Bo, J. Plasma miR-145 as a novel biomarker for the diagnosis and radiosensitivity prediction of human cervical cancer. J. Int. Med. Res. 2017, 45, 1054–1060. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Xiong, Y.; Li, Q. Interaction of lincRNA ROR and p53/miR-145 correlates with lung cancer stem cell signatures. J. Cell. Biochem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sheng, N.; Tan, G.; You, W.; Chen, H.; Gong, J.; Chen, D.; Zhang, H.; Wang, Z. miR-145 inhibits human colorectal cancer cell migration and invasion via PAK4-dependent pathway. Cancer Med. 2017, 6, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Hamano, R.; Miyata, H.; Yamasaki, M.; Kurokawa, Y.; Hara, J.; Moon, J.H.; Nakajima, K.; Takiguchi, S.; Fujiwara, Y.; Mori, M.; et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin. Cancer Res. 2011, 17, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.Q.; Xie, Z.C.; Tang, R.X.; Zhang, T.T.; Li, D.Y.; Li, Z.Y.; Chen, G. Clinical value of miR-145-5p in NSCLC and potential molecular mechanism exploration: A retrospective study based on GEO, qRT-PCR, and TCGA data. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.L.; Xu, L.Y.; Du, Z.P.; Liao, L.D.; Zhang, H.F.; Huang, Q.; Fang, G.Q.; Li, E.M. miRNA profile in esophageal squamous cell carcinoma: Downregulation of miR-143 and miR-145. World J. Gastroenterol. 2011, 17, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Liao, J.; Yang, M.; Sheng, J.; Yang, H.; Wang, Y.; Pan, E.; Guo, W.; Pu, Y.; Kim, S.J.; et al. The cluster of miR-143 and miR-145 affects the risk for esophageal squamous cell carcinoma through co-regulating fascin homolog 1. PLoS ONE 2012, 7, e33987. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.F.; Ma, X.Q.; Wang, L.P.; Wang, W. MicroRNA-145 exerts tumor-suppressive and chemo-resistance lowering effects by targeting CD44 in gastric cancer. World J. Gastroenterol. 2017, 23, 2337–2345. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhang, C.; Zhang, J.; Zhang, N.; Li, T.; Fang, J.; Zhang, Y.; Zuo, F.; Tao, Z.; Tang, S.; et al. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int. J. Oncol. 2017, 50, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Taniguchi, K.; Sugito, N.; Kuranaga, Y.; Inamoto, T.; Takahara, K.; Takai, T.; Yoshikawa, Y.; Kiyama, S.; Akao, Y.; et al. miR-145 negatively regulates Warburg effect by silencing KLF4 and PTBP1 in bladder cancer cells. Oncotarget 2017, 8, 33064–33077. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Cheng, J.; Zhang, J.; Zhang, Y.; Chen, X.; Xie, J.; Luo, S. miR-145 inhibits drug resistance to Oxaliplatin in colorectal cancer cells through regulating G protein coupled receptor 98. Zhonghua Wei Chang. Wai Ke Za Zhi 2017, 20, 566–570. [Google Scholar] [PubMed]
- Wang, T.Y.; Zhang, Q.Q.; Zhang, X.; Sun, Q.L.; Zhao, C.P.; Wang, X.Y. The effect of recombinant lentiviral vector encoding miR-145 on human esophageal cancer cells. Tumour Biol. 2015, 36, 9733–9738. [Google Scholar] [CrossRef] [PubMed]
- Kano, M.; Seki, N.; Kikkawa, N.; Fujimura, L.; Hoshino, I.; Akutsu, Y.; Chiyomaru, T.; Enokida, H.; Nakagawa, M.; Matsubara, H. miR-145, miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer 2010, 127, 2804–2814. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lu, J.; Ye, Z.; Han, X.; Zheng, X.; Hou, H.; Chen, W.; Li, X.; Zhao, L. 20(S)-Rg3 blocked epithelial-mesenchymal transition through DNMT3A/miR-145/FSCN1 in ovarian cancer. Oncotarget 2017. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, J.; Tao, L.; Zhang, K.; Wang, R.; Chu, X.; Chen, L. Long noncoding RNA ROR regulates chemoresistance in docetaxel-resistant lung adenocarcinoma cells via epithelial mesenchymal transition pathway. Oncotarget 2017, 8, 33144–33158. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Zhang, Y.; Tang, Y.; Li, Q. MALAT1 Modulates TGF-β1-Induced Endothelial-to-Mesenchymal Transition through Downregulation of miR-145. Cell. Physiol. Biochem. 2017, 42, 357–372. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.E.; Du, P.Z.; Zhang, H.D.; Huang, G. Long Noncoding RNA CRNDE Promotes Proliferation of Gastric Cancer Cells by Targeting miR-145. Cell. Physiol. Biochem. 2017, 42, 13–21. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mei, L.-L.; Wang, W.-J.; Qiu, Y.-T.; Xie, X.-F.; Bai, J.; Shi, Z.-Z. miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma. Int. J. Mol. Sci. 2017, 18, 1833. https://doi.org/10.3390/ijms18091833
Mei L-L, Wang W-J, Qiu Y-T, Xie X-F, Bai J, Shi Z-Z. miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma. International Journal of Molecular Sciences. 2017; 18(9):1833. https://doi.org/10.3390/ijms18091833
Chicago/Turabian StyleMei, Li-Li, Wen-Jun Wang, Yun-Tan Qiu, Xiu-Feng Xie, Jie Bai, and Zhi-Zhou Shi. 2017. "miR-145-5p Suppresses Tumor Cell Migration, Invasion and Epithelial to Mesenchymal Transition by Regulating the Sp1/NF-κB Signaling Pathway in Esophageal Squamous Cell Carcinoma" International Journal of Molecular Sciences 18, no. 9: 1833. https://doi.org/10.3390/ijms18091833