Genome-Wide Identification and Characterization of the GmSnRK2 Family in Soybean
Abstract
:1. Introduction
2. Results
2.1. Identification and Properties of SnRK2 Genes in Soybean
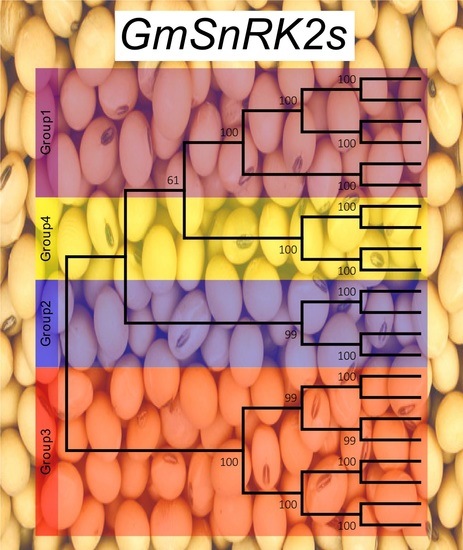
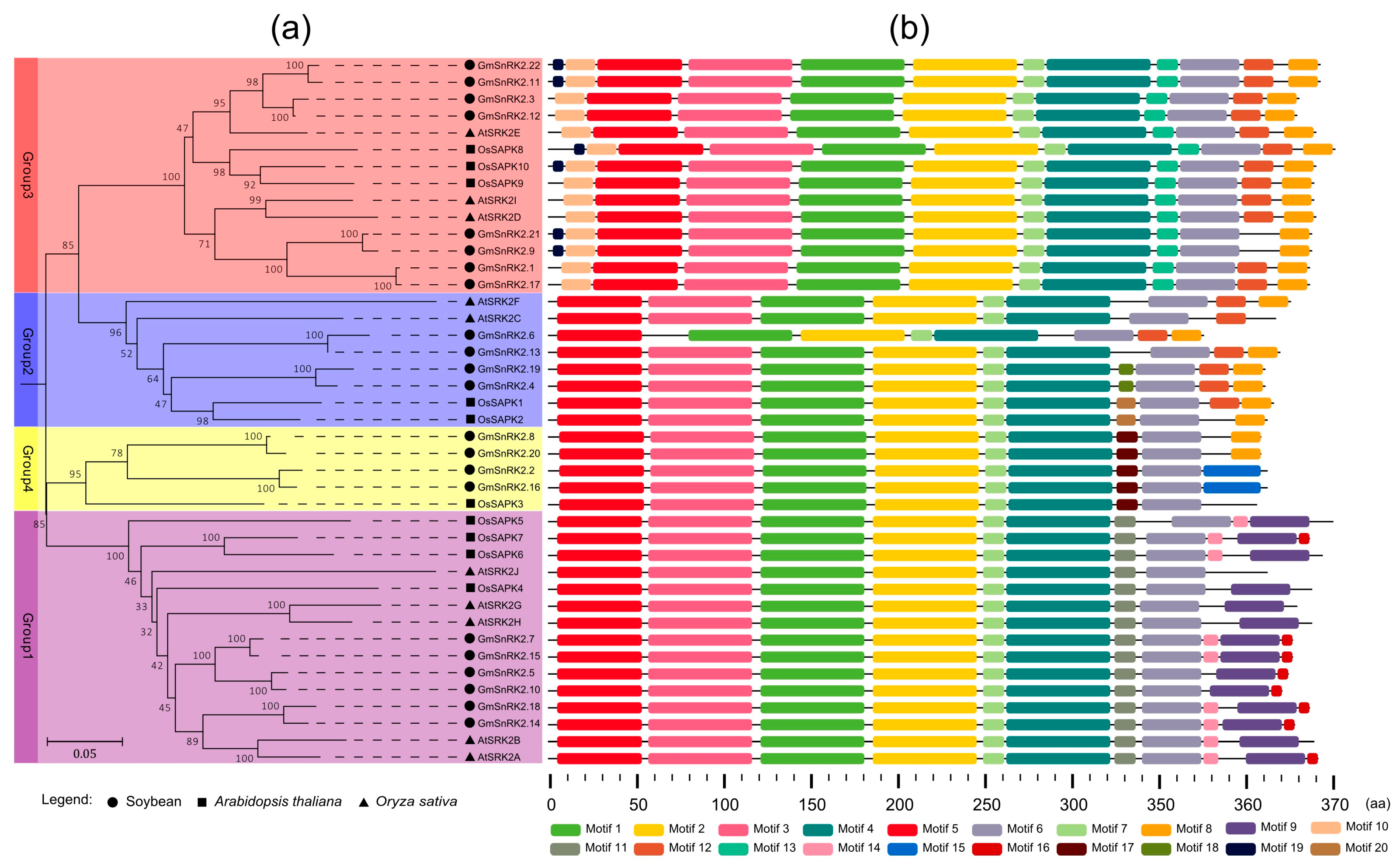
2.2. Phylogenetic and Motif Analysis of SnRK2 Genes in Soybean
2.3. Cis-Acting Regulatory Elements in the Promoters of GmSnRK2 Genes
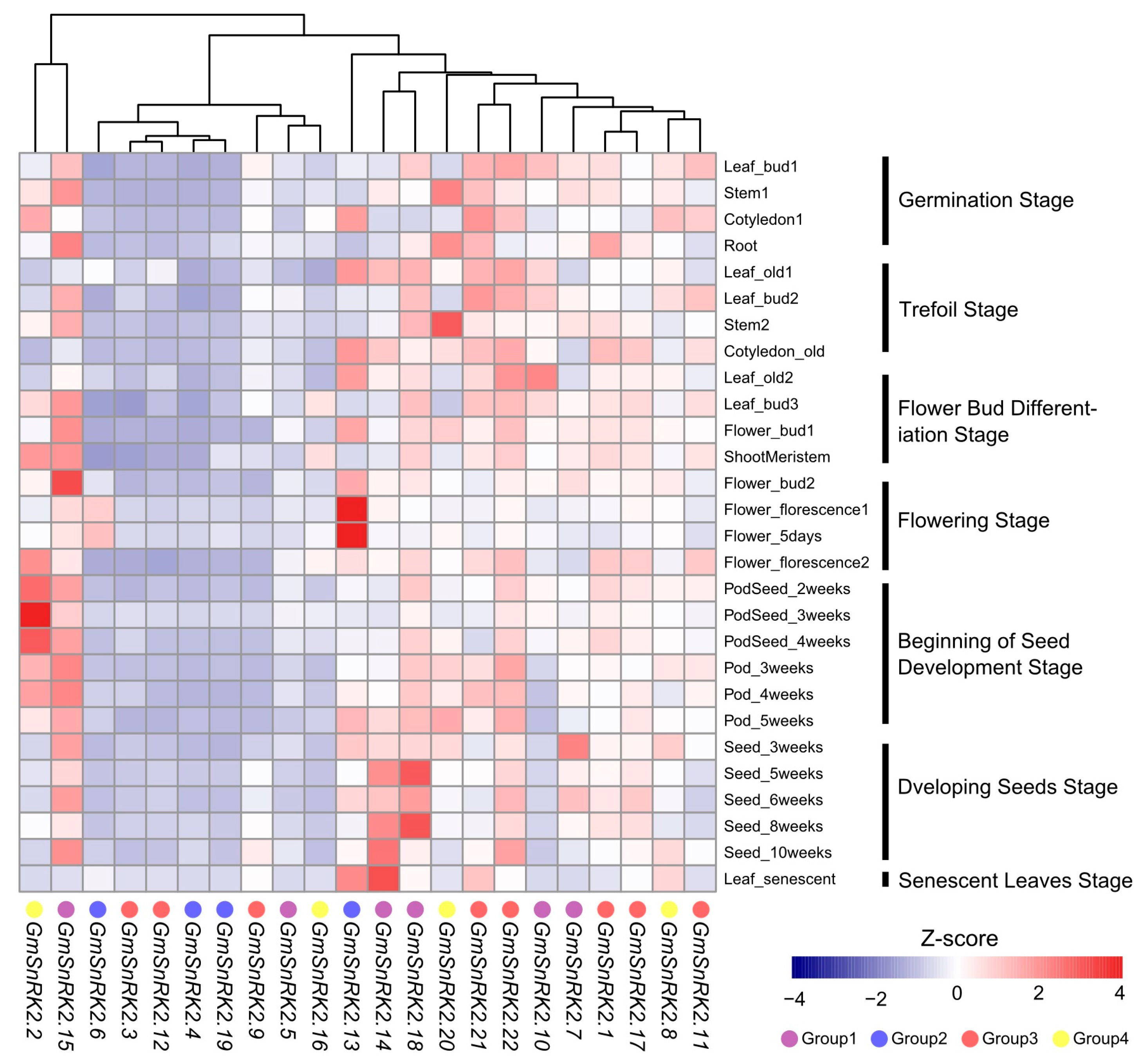
2.4. Expression Analysis of GmSnRK2 Genes during Growth and Development of Soybean
2.5. Differential Expression of GmSnRK2s in Response to Salt Stress, Abscisic Acid and Strigolactones
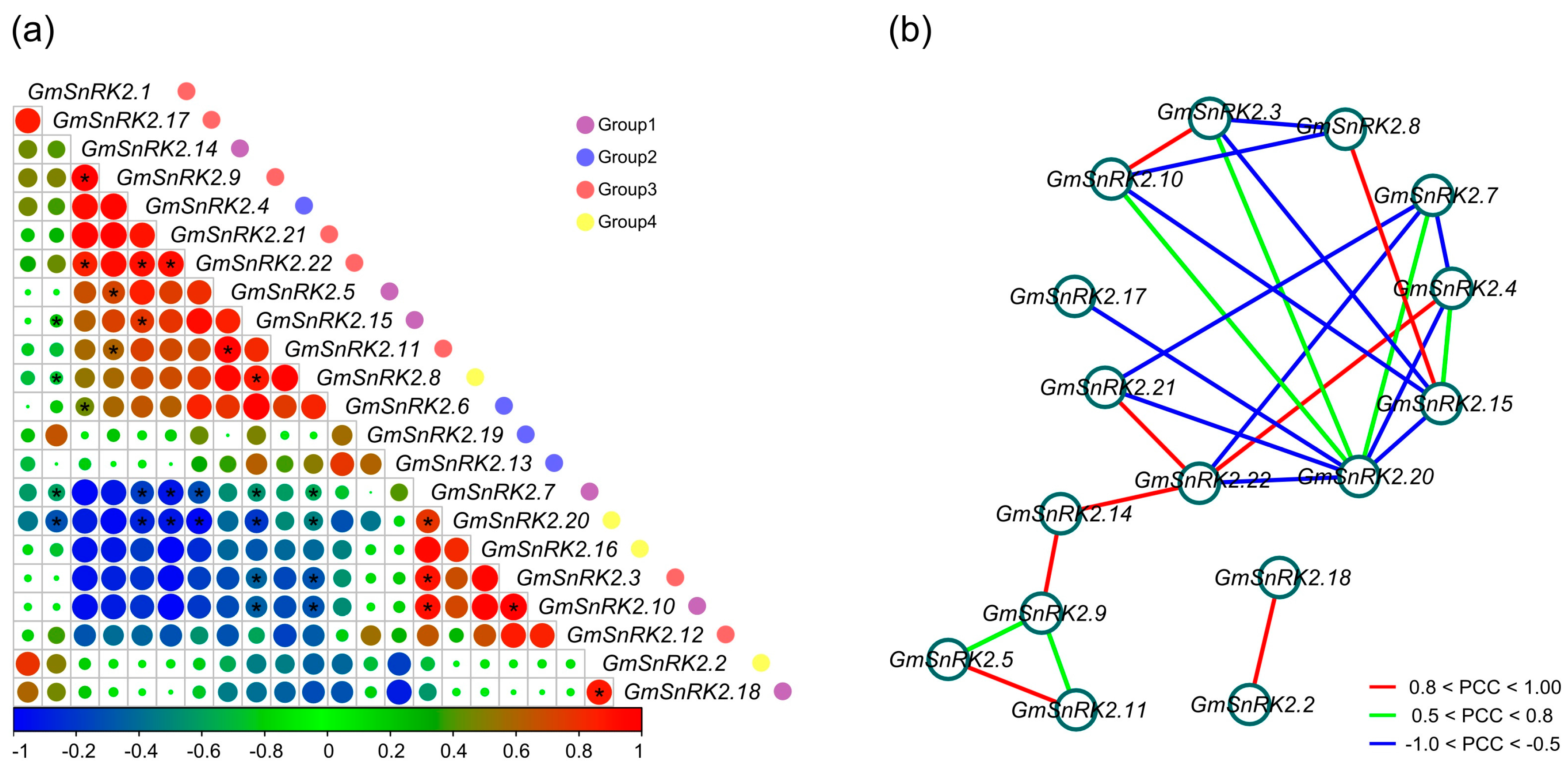
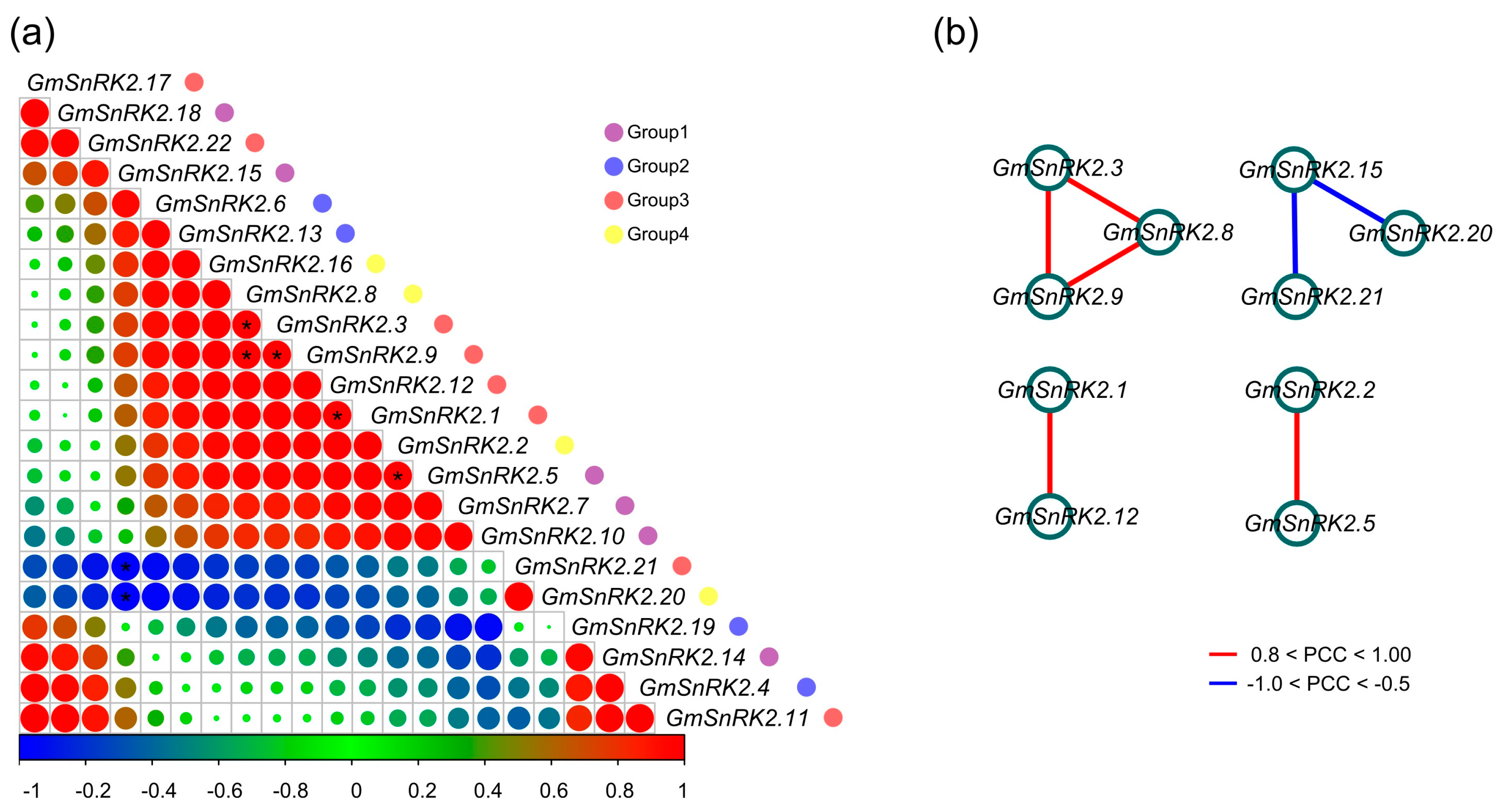
2.6. Co-Regulatory Networks of GmSnRK2s
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Identification of GmSnRK2 Genes and Gene Structure Analysis
4.3. Functional Domain and Cis-Acting Regulatory Elements Analysis
4.4. Multiple Sequence Alignment and Phylogenetic Analysis
4.5. Expression Analysis
4.6. RNA Extraction and cDNA Synthesis
4.7. Quantitative Real-Time Polymerase Chain Reaction
4.8. Pearson Correlation Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| SLs | Strigolactones |
| MeJA | Methyl Jasmonic acid |
| ERE | Ethylene response element |
| JGI | Joint Genome Institute |
| LTR | Low-temperature responsiveness |
| MBS | MYB binding site involved in drought-inducibility |
| GARE | Gibberellin response element |
| HSE | Heat stress responsiveness |
| WUN-motif | Wound-responsive element |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| FPKM | Fragments Per Kilobase Of Exon Per Million Fragments Mapped |
| GRAVY | Grand average of hydropathy |
| PCCs | Pearson correlation coefficients |
| CDS | Coding sequence |
| TSS | Transcription start site |
| MW | Molecular weight |
| pI | Isoelectric point |
| aa | Amino acid |
| bp | Base pair |
| kDa | Kilodalton |
References
- Bohnert, H.J.; Gong, Q.; Li, P.; Ma, S. Unraveling abiotic stress tolerance mechanisms–Getting genomics going. Curr. Opin. Plant Biol. 2006, 9, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 2006, 9, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Gibberellins and abscisic acid signal crosstalk: Living and developing under unfavorable conditions. Plant Cell Rep. 2013, 32, 1007–1016. [Google Scholar] [CrossRef] [PubMed]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z.K.; Sakuma, Y.; Furihata, T.; Abe, H.; Narusaka, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabfidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Jia, W.; Yang, J.C.; Ismail, A.M. Role of ABA in integrating plant responses to drought and salt stresses. Field Crop Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Klingler, J.P.; Batelli, G.; Zhu, J.K. ABA receptors: The START of a new paradigm in phytohormone signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef] [PubMed]
- Furihata, T.; Maruyama, K.; Fujita, Y.; Umezawa, T.; Yoshida, R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA 2006, 103, 1988–1993. [Google Scholar] [CrossRef] [PubMed]
- Ghillebert, R.; Swinnen, E.; Wen, J.; Vandesteene, L.; Ramon, M.; Norga, K.; Rolland, F.; Winderickx, J. The AMPK/SNF1/SnRK1 fuel gauge and energy regulator: Structure, function and regulation. FEBS J. 2011, 278, 3978–3990. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Hobo, T.; Ichimura, K.; Mizoguchi, T.; Takahashi, F.; Aronso, J.; Ecker, J.R.; Shinozaki, K. ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 2002, 43, 1473–1483. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Yamamoto, S.; Minami, H.; Kagaya, Y.; Hattori, T. Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 2004, 16, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Boudsocq, M.; Barbier-Brygoo, H.; Laurière, C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 2004, 279, 41758–41766. [Google Scholar] [CrossRef] [PubMed]
- Saha, J.; Chatterjee, C.; Sengupta, A.; Gupta, K.; Gupta, B. Genome-wide analysis and evolutionary study of sucrose non-fermenting 1-related protein kinase 2 (SnRK2) gene family members in Arabidopsis and Oryza. Comput. Biol. Chem. 2014, 49, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Qin, Y.; Zou, Y.; Ma, F. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene 2014, 552, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Huai, J.; Wang, M.; He, J.; Zheng, J.; Dong, Z.; Lv, H.; Zhao, J.; Wang, G. Cloning and characterization of the SnRK2 gene family from Zea mays. Plant Cell Rep. 2008, 27, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Mao, X.; Zhang, H.; Chen, S.; Zhai, C.; Yang, S.; Jing, R. Cloning and characterization of TaSnRK2.3, a novel SnRK2 gene in common wheat. J. Exp. Bot. 2013, 64, 2063–2080. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mao, X.; Jing, R.; Chang, X.; Xie, H. Characterization of a common wheat (Triticum aestivum L.) TaSnRK2.7 gene involved in abiotic stress responses. J. Exp. Bot. 2011, 62, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Tian, S.; Chang, X.; Jing, R. TaSnRK2.4, a SNF1-type serine-threonine protein kinase of wheat (Triticum aestivum L.) confers enhanced multi-stress tolerance in Arabidopsis. J. Exp. Bot. 2009, 61, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Mao, X.; Wang, C.; Jing, R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS ONE 2010, 5, e16041. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.J.; Ma, T.; Zhu, N.; Liu, L.; Harmon, A.C.; Wang, Q.; Chen, S. Genome-wide identification and homeolog-specific expression analysis of the SnRK2 genes in Brassica napus guard cells. Plant Mol. Biol. 2016, 91, 211–227. [Google Scholar] [CrossRef] [PubMed]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tang, J.; Duan, W.; Wang, Z.; Song, X.; Hou, X. Molecular evolution, characterization, and expression analysis of SnRK2 gene family in Pak-choi (Brassica rapa ssp. chinensis). Front. Plant Sci. 2015, 6, 879. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ge, X.; Yang, Z.; Zhang, C.; Zhao, G.; Chen, E.; Liu, J.; Zhang, X.; Li, F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 2017, 18, 54. [Google Scholar] [CrossRef] [PubMed]
- Kulik, A.; Wawer, I.; Krzywinska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein kinases—Key regulators of plant response to abiotic stresses. OMICS 2011, 15, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Soon, F.F.; Ng, L.M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Fujita, Y.; Kanamori, N.; Katagiri, T.; Umezawa, T.; Kidokoro, S.; Maruyama, K.; Yoshida, T.; Ishiyama, K.; Kobayashi, M.; et al. Three arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009, 50, 1345–1363. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Takahashi, F.; Anderson, J.C.; Ishihama, Y.; Peck, S.C.; Shinozaki, K. Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 2013, 6, rs8. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The regulatory domain of SRK2E/OST1/SnRK2.6 interacts with ABI1 and integrates abscisic acid (ABA) and osmotic stress signals controlling stomatal closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Yoshida, R.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-response gene expression in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2004, 101, 17306–17311. [Google Scholar] [CrossRef] [PubMed]
- Chae, M.J.; Lee, J.S.; Nam, M.H.; Cho, K.; Hong, J.Y.; Yi, S.A.; Suh, S.C.; Yoon, I.S. A rice dehydration-inducible SNF1-related protein kinase 2 phosphorylates an abscisic acid responsive element-binding factor and associates with ABA signaling. Plant Mol. Biol. 2007, 63, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.W.; Kim, M.C.; Shin, P.G.; Kim, J.S.; Kim, C.Y.; Lee, S.Y.; Hwang, I.; Bahk, J.D.; Hong, J.C.; Han, C.; et al. Differential expression of two functional serine/threonine protein kinases from soybean that have an unusual acidic domain at the carboxy terminus. Mol. Gen. Genet. 1997, 255, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Monks, D.E.; Aghoram, K.; Courtney, P.D.; DeWald, D.B.; Dewey, R.E. Hyperosmotic stress induces the rapid phosphorylation of a soybean phosphatidylinositol transfer protein homolog through activation of the protein kinases SPK1 and SPK2. Plant Cell 2001, 13, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Nakashima, K.; Miyakawa, T.; Kuromori, T.; Tanokura, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 2010, 51, 1821–1839. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K. Osmotic stress signaling via protein kinases. Cell Mol. Life Sci. 2012, 69, 3165–3173. [Google Scholar] [CrossRef] [PubMed]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.B.; Wu, F.Q.; Sheng, P.K.; Zhang, Z.; Zhang, X.; Guo, X.P.; Wang, J.L.; Cheng, Z.J.; Wang, J.; Wang, H.Y.; et al. The SnRK2-APC/CTE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 2015, 6, 7981. [Google Scholar] [CrossRef] [PubMed]
- Park, S-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Hotta, H.; Aoki, N.; Matsuda, T.; Adachi, T. Molecular analysis of a novel protein kinase in maturing rice seed. Gene 1998, 213, 47–54. [Google Scholar] [CrossRef]
- Xu, M.R.; Huang, L.Y.; Zhang, F.; Zhu, L.H.; Zhou, Y.L.; Li, Z.K. Genome-wide phylogenetic analysis of stress-activated protein kinase genes in rice (OsSAPKs) and expression profiling in response to Xanthomonas oryzae pv. oryzicola infection. Plant Mol. Biol. Rep. 2013, 31, 877. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant 2013, 147, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Verslues, P.E.; Zhu, J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 2007, 19, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Xu, X.; Crosley, R.A.; Greenwalt, S.A.; Sun, Y.; Blakeslee, B.; Wang, L.; Ni, W.; Sopko, M.S.; Yao, C.; et al. The protein kinase SnRK2.6 mediates the regulation of sucrose metabolism and plant growth in Arabidopsis. Plant Physiol. 2010, 153, 99–113. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.; Davidson, E.H. Gene regulatory networks for development. Proc. Natl. Acad. Sci. USA 2005, 102, 4936–4942. [Google Scholar] [CrossRef] [PubMed]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhou, Z.; Wang, Z.; Li, W.; Fang, C.; Wu, M.; Ma, Y.; Liu, T.; Kong, L.A.; Peng, D.L.; et al. Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 2014, 26, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
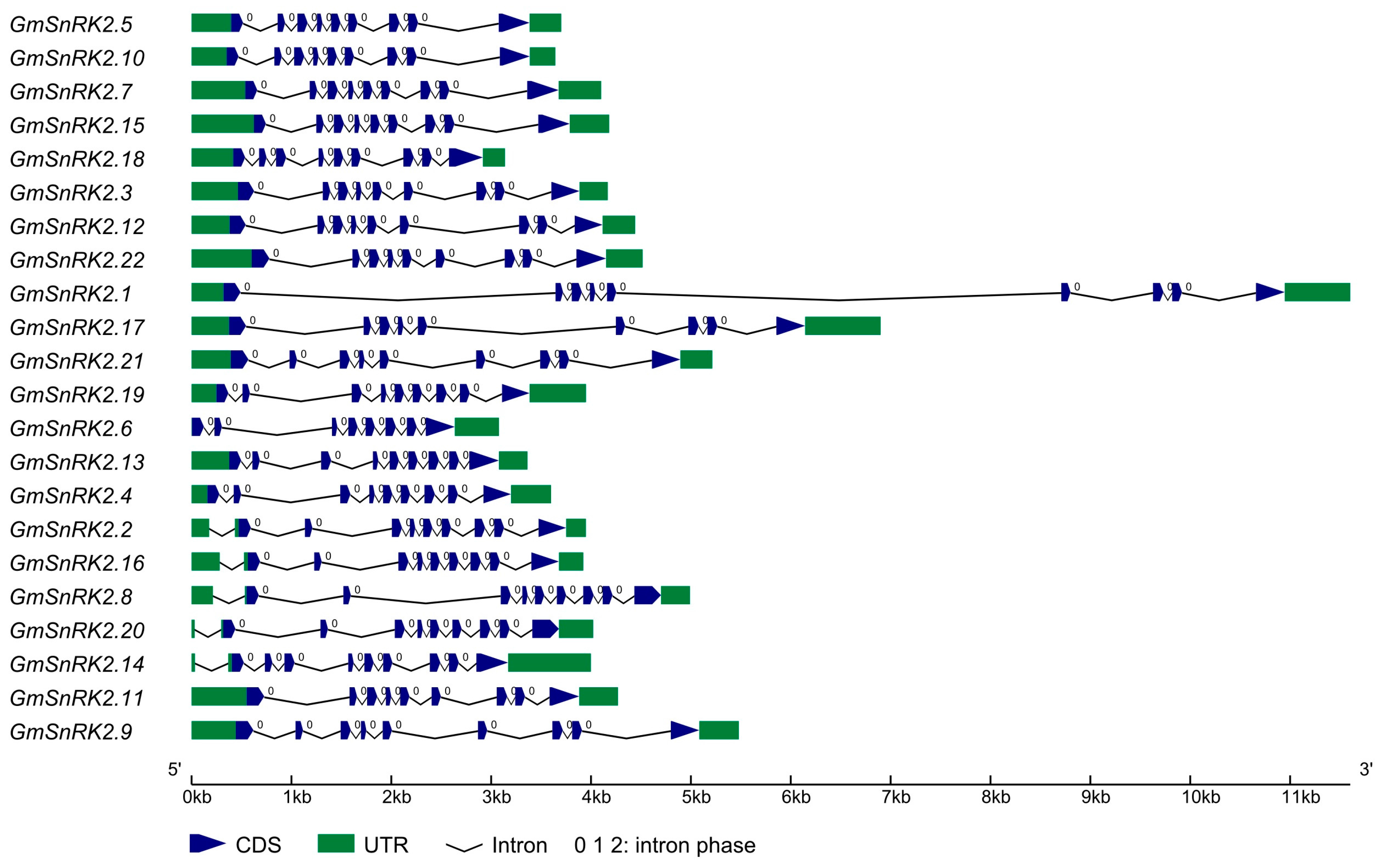

| Gene Name | Locus Name | Gene Location 1 | Transcript Length (bp) | Protein Length (aa) | Molecular Weight (kDa) | Isoelectric Point (pI) | Grand Average of Hydropathy (GRAVY) | Best Hits of Arabidopsis and Rice | Domain (Start–End aa) 2 |
|---|---|---|---|---|---|---|---|---|---|
| GmSnRK2.1 | Glyma01g39020 | Chr01:50978690–50990291: − | 2056 | 359 | 40.83 | 4.69 | −0.245 | AtSRK2I; OsSAPK10 | IPR017441 (27–50); IPR008271 (136–148) |
| GmSnRK2.2 | Glyma01g41260 (SPK1) | Chr01:52822191–52826139: + | 1432 | 339 | 38.56 | 6.44 | −0.432 | AtSRK2E; OsSAPK3 | IPR017441 (11–34); IPR008271 (120–132) |
| GmSnRK2.3 | Glyma02g15330 | Chr02:13828174–13832341: + | 1813 | 354 | 40.21 | 5.04 | −0.359 | AtSRK2E; OsSAPK10 | IPR017441 (24–47); IPR008271 (133–145) |
| GmSnRK2.4 | Glyma02g37090 | Chr02:42422548–42426147: − | 1577 | 338 | 38.3 | 5.38 | −0.267 | AtSRK2C; OsSAPK2 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.5 | Glyma04g38270 (SPK4) | Chr04:44680353–44684054: + | 1763 | 349 | 40.35 | 6.1 | −0.497 | AtSRK2A; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.6 | Glyma05g31000 | Chr05:36237913–36240990: + | 1376 | 309 | 35.37 | 5.17 | −0.366 | AtSRK2C; OsSAPK2 | IPR017441 (10–33); IPR008271 (85–97) |
| GmSnRK2.7 | Glyma05g33170 | Chr05:37897658–37901759: − | 2020 | 351 | 40.43 | 5.95 | −0.523 | AtSRK2A; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.8 | Glyma05g05540 | Chr05:4967484–4972474: − | 1532 | 336 | 38.12 | 5.6 | −0.447 | AtSRK2E; OsSAPK3 | IPR017441 (11–34); IPR008271 (120–132) |
| GmSnRK2.9 | Glyma05g09460 | Chr05:9361881–9367360: − | 1922 | 360 | 40.8 | 4.75 | −0.268 | AtSRK2I; OsSAPK10 | IPR017441 (29–52); IPR008271 (138–150) |
| GmSnRK2.10 | Glyma06g16780 | Chr06:13183468–13187110: − | 1648 | 346 | 39.98 | 6.48 | −0.468 | AtSRK2A; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.11 | Glyma07g29500 | Chr07:34352503–34356772: − | 2034 | 364 | 41.46 | 4.93 | −0.334 | AtSRK2E; OsSAPK10 | IPR017441 (29–52); IPR008271 (138–150) |
| GmSnRK2.12 | Glyma07g33120 | Chr07:38032643–38037084: − | 1769 | 353 | 40.07 | 5.04 | −0.344 | AtSRK2E; OsSAPK10 | IPR017441 (24–47); IPR008271 (133–145) |
| GmSnRK2.13 | Glyma08g14210 | Chr08:10340382–10343745: + | 1701 | 345 | 39.24 | 5.15 | −0.3 | AtSRK2E; OsSAPK1 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.14 | Glyma08g20090 | Chr08:15173104–15177102: − | 1959 | 352 | 40.26 | 6.1 | −0.486 | AtSRK2B; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.15 | Glyma08g00770 (SPK3) | Chr08:400210–404391: − | 2076 | 351 | 40.39 | 5.75 | −0.529 | AtSRK2A; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.16 | Glyma11g04150 (SPK2) | Chr11:2759687–2763609: − | 1585 | 339 | 38.66 | 6.39 | −0.45 | AtSRK2E; OsSAPK3 | IPR017441 (11–34); IPR008271 (120–132) |
| GmSnRK2.17 | Glyma11g06250 | Chr11:4436484–4443383: + | 2211 | 359 | 40.78 | 4.68 | −0.234 | AtSRK2I; OsSAPK10 | IPR017441 (27–50); IPR008271 (136–148) |
| GmSnRK2.18 | Glyma12g29130 | Chr12:32505082–32508220: + | 1719 | 359 | 41.08 | 5.82 | −0.537 | AtSRK2B; OsSAPK7 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.19 | Glyma14g35380 | Chr14:44313521–44317470: − | 1831 | 338 | 38.18 | 5.38 | −0.251 | AtSRK2C; OsSAPK1 | IPR017441 (10–33); IPR008271 (119–131) |
| GmSnRK2.20 | Glyma17g15860 | Chr17:12585658–12589679: − | 1405 | 336 | 38.04 | 5.71 | −0.46 | AtSRK2E; OsSAPK3 | IPR017441 (11–34); IPR008271 (120–132) |
| GmSnRK2.21 | Glyma17g20610 | Chr17:19751156–19756370: + | 1796 | 360 | 40.91 | 4.75 | −0.273 | AtSRK2I; OsSAPK10 | IPR017441 (29–52); IPR008271 (138–150) |
| GmSnRK2.22 | Glyma20g01240 | Chr20:846957–851473: − | 2064 | 364 | 41.49 | 4.89 | −0.344 | AtSRK2E; OsSAPK10 | IPR017441 (29–52); IPR008271 (138–150) |
| Group | Gene Name | Cis Elements | Number | Sequence Pattern (5′-3′) | Position (Strand) | Function |
|---|---|---|---|---|---|---|
| Group 1 | GmSnRK2.5 | CGTCA-motif | 1 | CGTCA | −410 bp (−) | MeJA Response Element |
| ERE | 3 | ATTTCAAA | −526 bp (−); −911 bp (+); −750 bp (+) | Ethylene Response element | ||
| P-box | 2 | CCTTTTG; GCCTTTTGAGT | −68 bp (−); −308 bp (−) | Gibberellin Response Element | ||
| TGACG-motif | 1 | TGACG | −410 bp (+) | MeJA Response Element | ||
| GmSnRK2.7 | CGTCA-motif | 2 | CGTCA | −717 bp (−); −834 bp (−) | MeJA Response Element | |
| TGACG-motif | 2 | TGACG | −717 bp (+); −834 bp (+) | MeJA Response Element | ||
| GARE-motif | 1 | TCTGTTG | −293 bp (−) | Gibberellin Response Element | ||
| TCA-element | 1 | CCATCTTTTT | −1412 bp (−) | Salicylic acid Response Element | ||
| TGA-element | 2 | AACGAC | −836 bp (−); −1438 bp (+) | Auxin Response Element | ||
| GmSnRK2.10 | ERE | 1 | ATTTCAAA | −359 bp (−) | Ethylene Response element | |
| GmSnRK2.14 | ERE | 1 | ATTTCAAA | −211 bp (+) | Ethylene Response element | |
| TATC-box | 1 | TATCCCA | −1289 bp (−) | Gibberellin Response Element | ||
| GmSnRK2.15 | AuxRR-core | 1 | GGTCCAT | −1409 bp (−) | Auxin Response Element | |
| ERE | 1 | ATTTCAAA | −972 bp (−) | Ethylene Response element | ||
| TGA-element | 1 | AACGAC | −559 bp (+) | Auxin Response Element | ||
| GmSnRK2.18 | ABRE | 1 | TACGTG | −516 bp (+) | ABA Response Element | |
| CGTCA-motif | 1 | CGTCA | −1228 bp (−) | MeJA Response Element | ||
| TGACG-motif | 1 | TGACG | −1228 bp (+) | MeJA Response Element | ||
| ERE | 2 | ATTTCAAA | −394 bp (+); −628 bp (+) | Ethylene Response element | ||
| TCA-element | 2 | GAGAAGAATA; CCATCTTTTT | −325 bp (−); −377 bp (+) | Salicylic acid Response Element | ||
| TGA-element | 1 | AACGAC | −837 bp (+) | Auxin Response Element | ||
| Group 2 | GmSnRK2.4 | ABRE | 1 | CACGTG | −1258 bp (−) | ABA Response Element |
| GARE-motif | 1 | AAACAGA | −1347 bp (−) | Gibberellin Response Element | ||
| GmSnRK2.6 | GARE-motif | 1 | AAACAGA | −314 bp (+) | Gibberellin Response Element | |
| TCA-element | 3 | CAGAAAAGGA; CCATCTTTTT; CAGAAAAGGA | −317 bp (+); −1352 bp (−); −564 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.13 | ABRE | 2 | GACACGTGGC; TACGTG | −657 bp (−); −1212 bp (−) | ABA Response Element | |
| ERE | 2 | ATTTCAAA | −99 bp (−); −1336 bp (+) | Ethylene Response element | ||
| P-box | 1 | GCCTTTTGAGT | −510 bp (+) | Gibberellin Response Element | ||
| TGA-element | 1 | AACGAC | −84 bp (+) | Auxin Response Element | ||
| GmSnRK2.19 | ABRE | 1 | CACGTG | −1401 bp (−) | ABA Response Element | |
| GARE-motif | 1 | AAACAGA | −1483 bp (−) | Gibberellin Response Element | ||
| Group 3 | GmSnRK2.1 | ABRE | 1 | TACGTG | −79 bp (+) | ABA Response Element |
| AuxRR-core | 1 | GGTCCAT | −1158 bp (−) | Auxin Response Element | ||
| P-box | 1 | CCTTTTG | −115 bp (−) | Gibberellin Response Element | ||
| TCA-element | 2 | CCATCTTTTT; GAGAAGAATA | −164 bp (−); −412 bp (−) | Salicylic acid Response Element | ||
| GmSnRK2.3 | CGTCA-motif | 3 | CGTCA | −261 bp (+); −1314 bp (+); −295 bp (+) | MeJA Response Element | |
| TGACG-motif | 3 | TGACG | −261 bp (−); −1314 bp (−); −295 bp (−) | MeJA Response Element | ||
| ERE | 1 | ATTTCAAA | −481 bp (−) | Ethylene Response element | ||
| GARE-motif | 1 | AAACAGA | −62 bp (+) | Gibberellin Response Element | ||
| P-box | 1 | CCTTTTG | −1451 bp (−) | Gibberellin Response Element | ||
| TCA-element | 1 | GAGAAGAATA | −934 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.9 | ABRE | 1 | CCGCGTAGGC | −1360 bp (−) | ABA Response Element | |
| CGTCA-motif | 1 | CGTCA | −1386 bp (+) | MeJA Response Element | ||
| ERE | 1 | ATTTCAAA | −334 bp (−) | Ethylene Response element | ||
| GARE-motif | 1 | AAACAGA | −403 bp (−) | Gibberellin Response Element | ||
| GmSnRK2.11 | TCA-element | 2 | CCATCTTTTT | −1352 bp (+); −1469 bp (−) | Salicylic acid Response Element | |
| GmSnRK2.12 | ABRE | 2 | CACGTG; TACGTG | −538 bp (+); −1147 bp (−) | ABA Response Element | |
| AuxRR-core | 1 | GGTCCAT | −1345 bp (+) | Auxin Response Element | ||
| CGTCA-motif | 2 | CGTCA | −117 bp (+); −1361 bp (+) | MeJA Response Element | ||
| TGACG-motif | 2 | TGACG | −117 bp (−); −1361 bp (−) | MeJA Response Element | ||
| TCA-element | 2 | CCATCTTTTT | −98 bp (−); −710 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.17 | ABRE | 2 | CACGTG; AGTACGTGGC | −207 bp (+); −410 bp (−) | ABA Response Element | |
| AuxRR-core | 1 | GGTCCAT | −981 bp (−) | Auxin Response Element | ||
| ERE | 1 | ATTTCAAA | −371 bp (−) | Ethylene Response element | ||
| GARE-motif | 1 | AAACAGA | −126 bp (−) | Gibberellin Response Element | ||
| TATC-box | 1 | TATCCCA | −1305 bp (+) | Gibberellin Response Element | ||
| TCA-element | 1 | CCATCTTTTT | −780 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.21 | ABRE | 3 | CCTACGTGGC; ACGTGGC; TACGTG | −1349 bp (+); −1352 bp (+); −1351 bp (+) | ABA Response Element | |
| CGTCA-motif | 1 | CGTCA | −1374 bp (+) | MeJA Response Element | ||
| ERE | 1 | ATTTCAAA | −309 bp (−) | Ethylene Response element | ||
| TGACG-motif | 1 | TGACG | −1374 bp (−) | MeJA Response Element | ||
| GmSnRK2.22 | TGACG-motif | 1 | TGACG | −705 bp (−) | MeJA Response Element | |
| CGTCA-motif | 1 | CGTCA | −705 bp (+) | MeJA Response Element | ||
| TCA-element | 2 | GAGAAGAATA; TCAGAAGAGG | −425 bp (+); −792 bp (+) | Salicylic acid Response Element | ||
| Group 4 | GmSnRK2.2 | CGTCA-motif | 1 | CGTCA | −477 bp (−) | MeJA Response Element |
| TGACG-motif | 1 | TGACG | −477 bp (+) | MeJA Response Element | ||
| TCA-element | 1 | CCATCTTTTT | −82 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.16 | CGTCA-motif | 1 | CGTCA | −865 bp (−) | MeJA Response Element | |
| TGACG-motif | 1 | TGACG | −865 bp (+) | MeJA Response Element | ||
| TCA-element | 1 | CCATCTTTTT | −449 bp (+) | Salicylic acid Response Element | ||
| GmSnRK2.20 | GARE-motif | 1 | AAACAGA | −281 bp (+) | Gibberellin Response Element |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Cheng, Y.-H.; Zhang, C.; Shen, X.-J.; You, Q.-B.; Guo, W.; Li, X.; Song, X.-J.; Zhou, X.-A.; Jiao, Y.-Q. Genome-Wide Identification and Characterization of the GmSnRK2 Family in Soybean. Int. J. Mol. Sci. 2017, 18, 1834. https://doi.org/10.3390/ijms18091834
Zhao W, Cheng Y-H, Zhang C, Shen X-J, You Q-B, Guo W, Li X, Song X-J, Zhou X-A, Jiao Y-Q. Genome-Wide Identification and Characterization of the GmSnRK2 Family in Soybean. International Journal of Molecular Sciences. 2017; 18(9):1834. https://doi.org/10.3390/ijms18091834
Chicago/Turabian StyleZhao, Wei, Yi-Hui Cheng, Chi Zhang, Xin-Jie Shen, Qing-Bo You, Wei Guo, Xiang Li, Xue-Jiao Song, Xin-An Zhou, and Yong-Qing Jiao. 2017. "Genome-Wide Identification and Characterization of the GmSnRK2 Family in Soybean" International Journal of Molecular Sciences 18, no. 9: 1834. https://doi.org/10.3390/ijms18091834