Mitochondrial Biogenesis in Response to Chromium (VI) Toxicity in Human Liver Cells
Abstract
:1. Introduction
2. Results
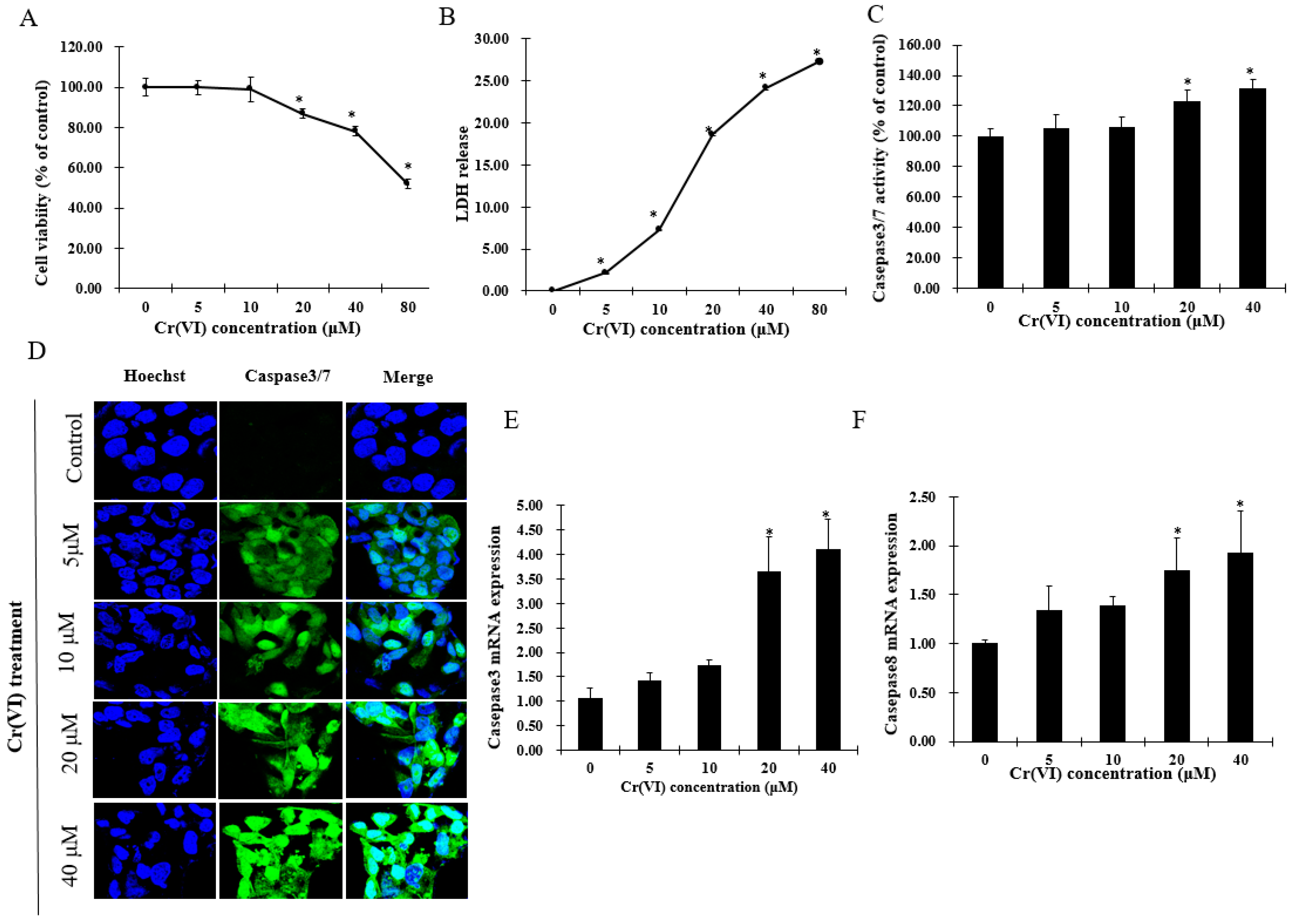
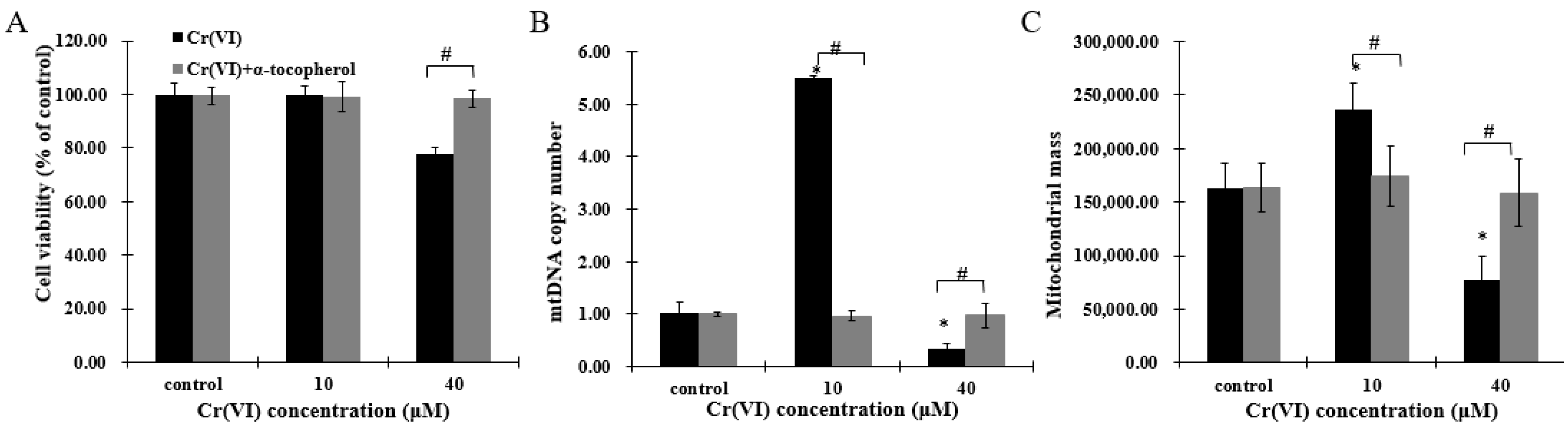
2.1. Cr(VI) Induces Cytotoxicity and Cell Death in HepG2 Cells
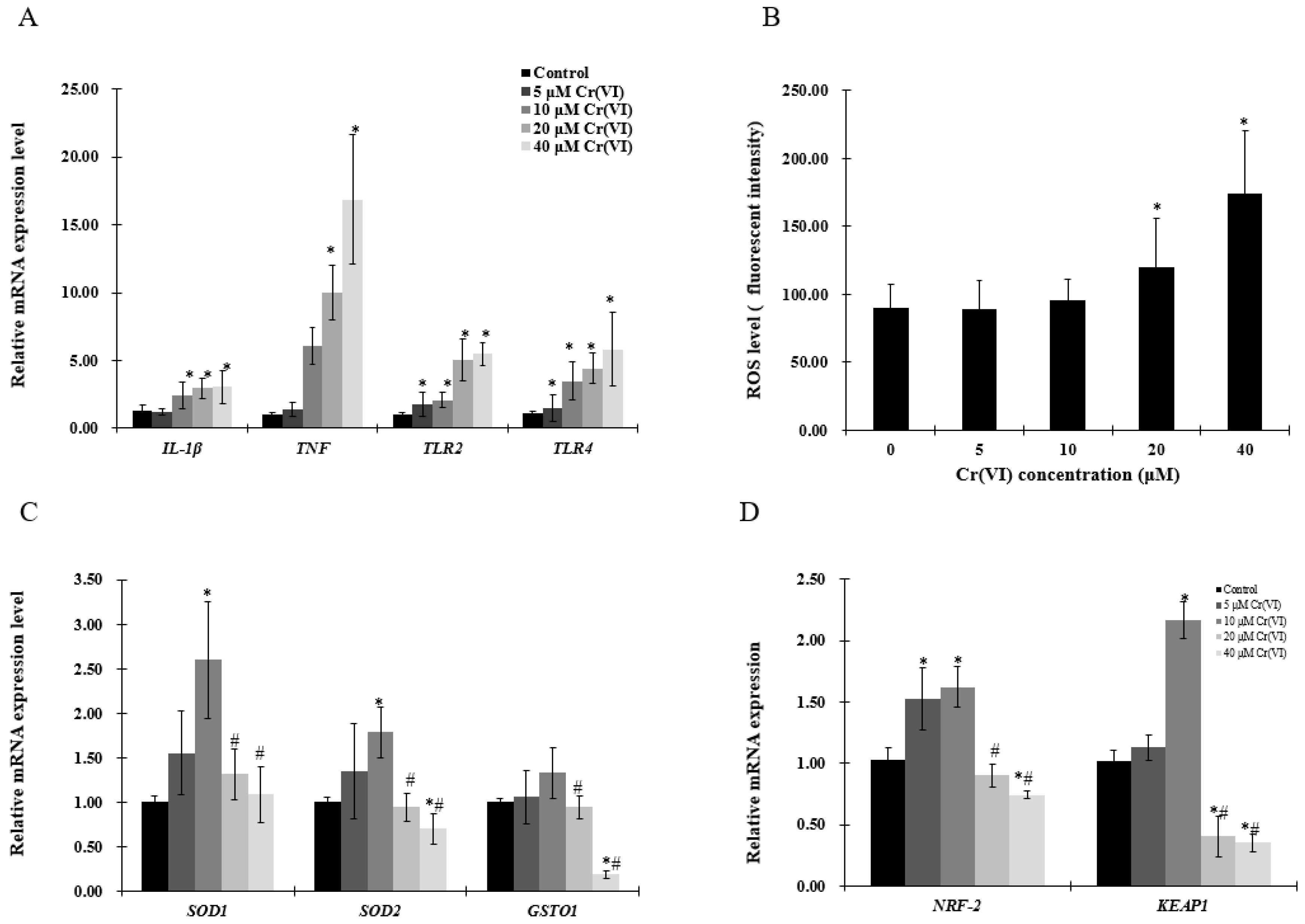
2.2. The Effect of Cr(VI) on the mRNA Level of Inflammatory Cytokines and Redox Reaction in HepG2 Cells
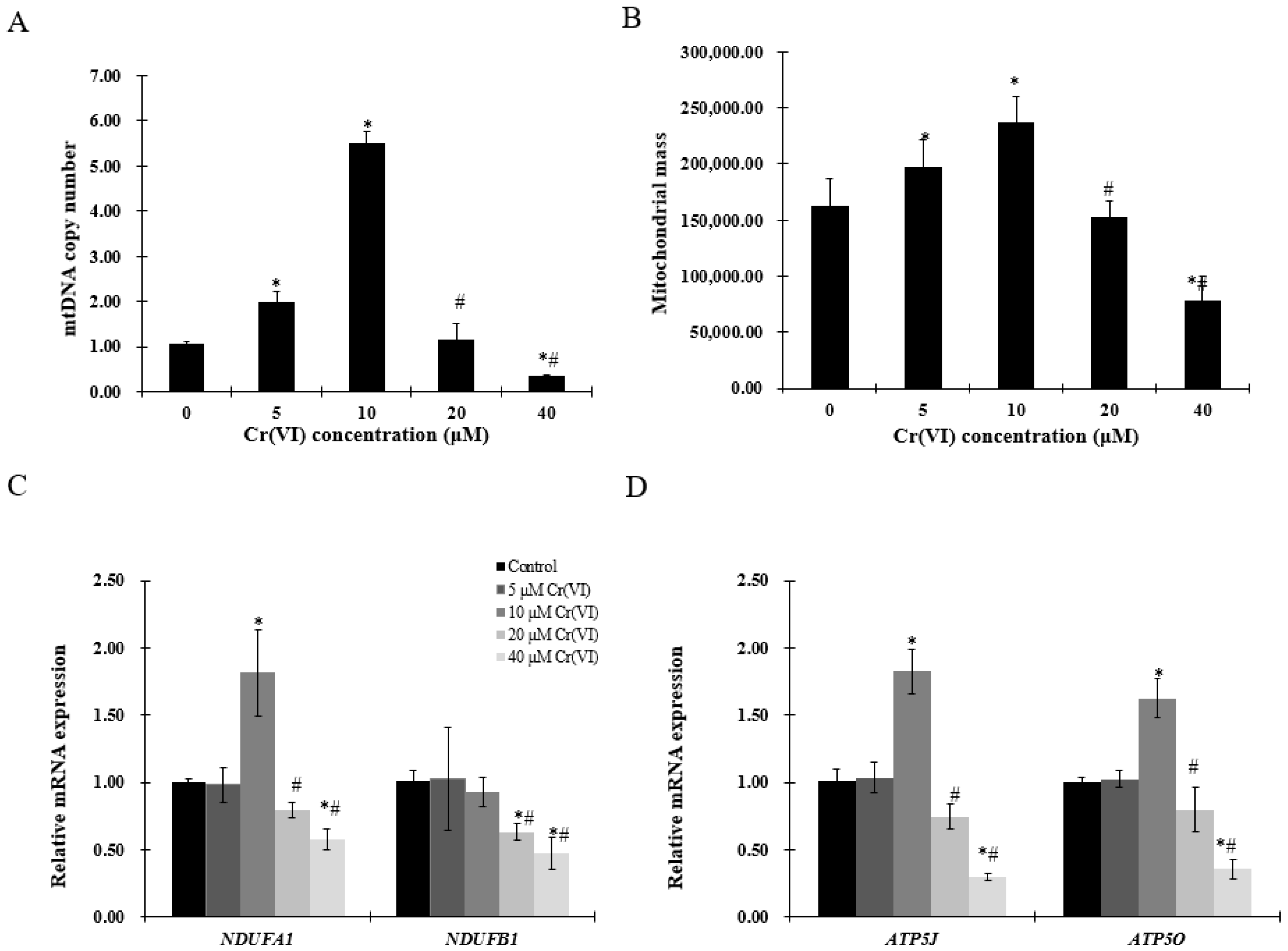
2.3. Exposure to Low Concentrations of Cr(VI) Induces Adaptive Upregulation of Mitochondrial Biogenesis
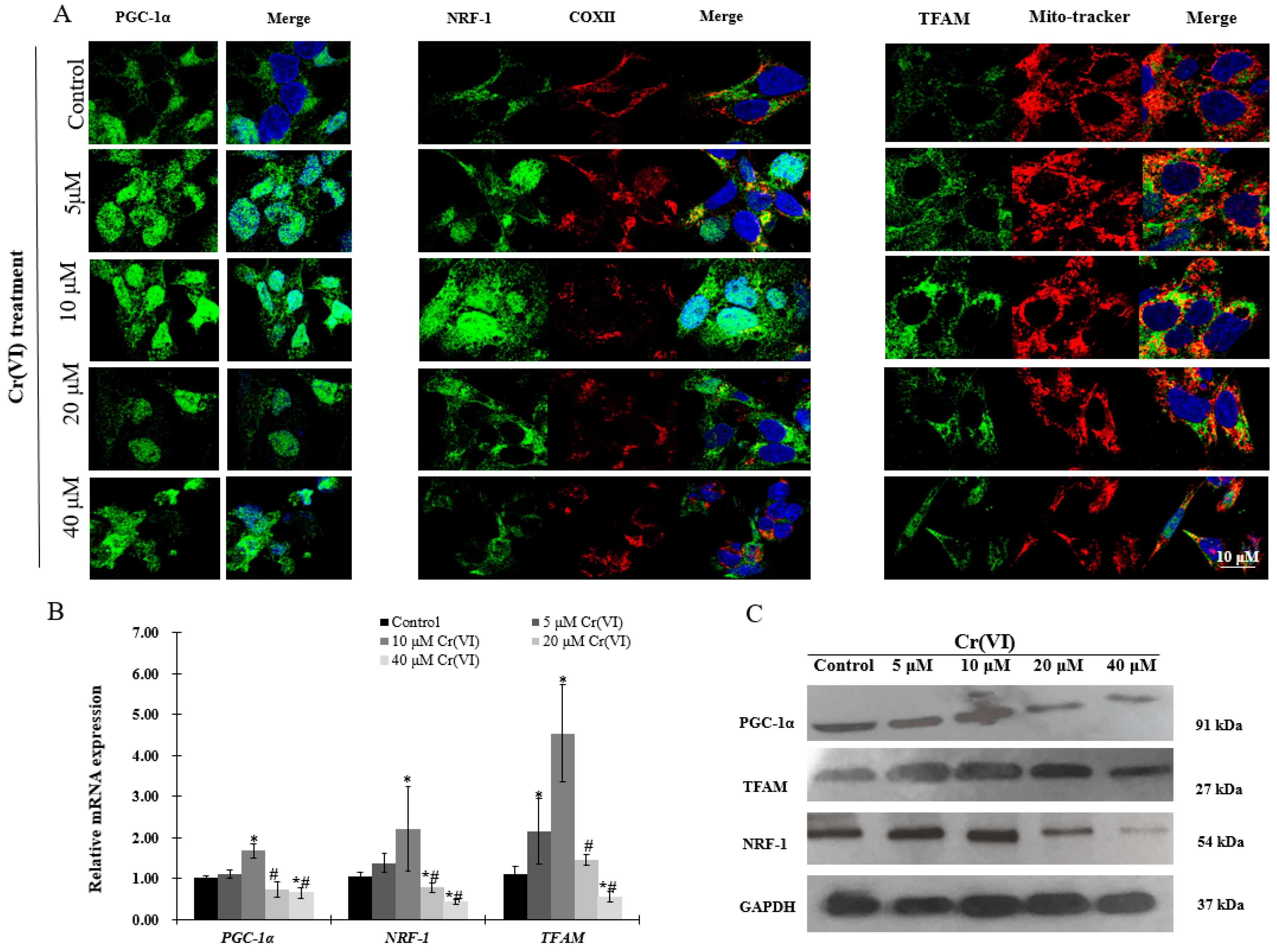
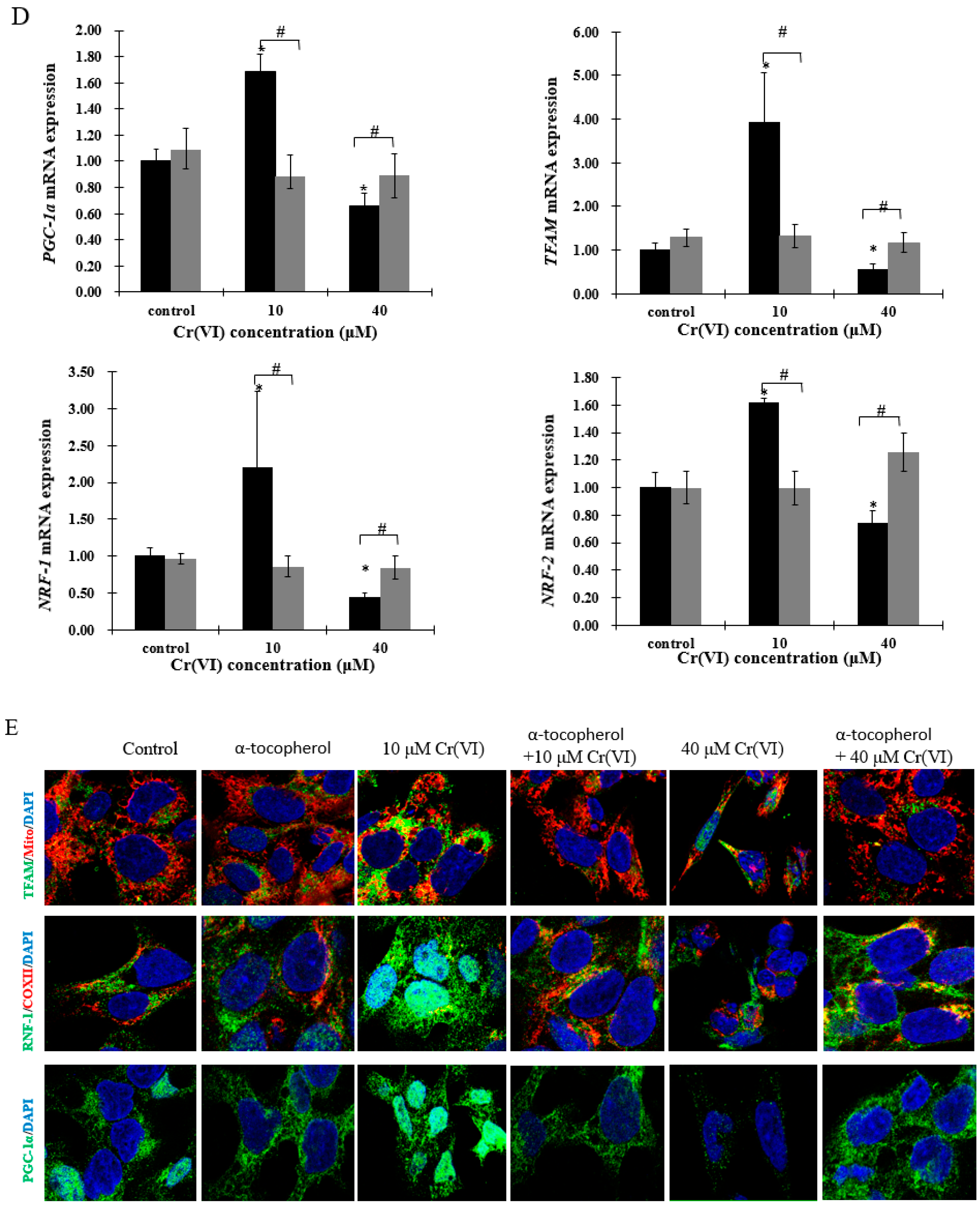
2.4. Upregulation of PGC-1α Expression and Its Downstream Targets at Low Concentrations of Cr(VI)
2.5. Expression of Genes Involved in Mitochondrial Biogenesis Pathway after Exposure to Cr(VI) in HepG2 Cells
2.6. The Effect of α-Tocopherol on Mitochondrial Biogenesis in HepG2 Cells Exposed to Cr(VI)
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. Treatment and Cytotoxicity Assay
4.4. Reactive Oxygen Species Measurement
4.5. RNA Extraction and Quantitative Real-Time qPCR
4.6. Mito-Tracker Assay
4.7. Measurement of Mitochondrial Mass
4.8. Immunofluorescence
4.9. Caspase-3/7 Staining
4.10. Western Blot Analysis
4.11. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AKT1 | Threonine kinase 1 |
| APT5O | ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| ATF2 | Transcription factor |
| ATP5J | ATP synthase, H+ transporting, mitochondrial Fo complex subunit F6 |
| Cr(VI) | Hexavalent chromium |
| CREB | cAMP response element-binding protein |
| FOXO1 | Forkhead box class-O |
| GSOT1 | Glutathione S-transferase omega 1 |
| HepG2 | Human Hepatocellular Carcinoma cells |
| IL-1β | Interleukin 1-β |
| KEAP1 | Kelch like ECH associate protein 1 |
| LDH | Lactate Dehydrogenase |
| MAPK1 | Mitogen-activated protein kinase 1 |
| mtDNA | Mitochondrial DNA |
| NDUFA1 | NADH: ubiquinone oxidoreductase subunit A1 |
| NDUFB1 | NADH: ubiquinone oxidoreductase subunit B1 |
| NRF | Nuclear respiratory factor |
| PGC-1α | Peroxisome-proliferator-activated receptor γ coactivator-1α |
| PTEN | Phosphatase and tensin homolog |
| ROS | Reactive oxygen species |
| SIRT1 | Silent information regulator-1 |
| SOD | Superoxide dismutase |
| TFAM | Mitochondrial transcription factor A |
| TLRs | Toll-like receptors |
| TNF | Tumor necrosis factor |
References
- Proctor, D.M.; Suh, M.; Mittal, L.; Hirsch, S.; Valdes Salgado, R.; Bartlett, C.; van Landingham, C.; Rohr, A.; Crump, K. Inhalation cancer risk assessment of hexavalent chromium based on updated mortality for painesville chromate production workers. J. Expo. Sci. Environ. Epidemiol. 2016, 26, 224–231. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Holmgren, S.; Andrews, D.M.; Wolfe, M.S. Evaluating the impact of the US National toxicology program: A case study on hexavalent chromium. Environ. Health Perspect. 2017, 125, 181–188. [Google Scholar] [PubMed]
- Stout, M.D.; Herbert, R.A.; Kissling, G.E.; Collins, B.J.; Travlos, G.S.; Witt, K.L.; Melnick, R.L.; Abdo, K.M.; Malarkey, D.E.; Hooth, M.J. Hexavalent chromium is carcinogenic to F344/N rats and B6C3F1 mice after chronic oral exposure. Environ. Health Perspect. 2009, 117, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Elshazly, M.O.; Morgan, A.M.; Ali, M.E.; Abdel-Mawla, E.; Abd El-Rahman, S.S. The mitigative effect of raphanus sativus oil on chromium-induced geno- and hepatotoxicity in male rats. J. Adv. Res. 2016, 7, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Bosgelmez, I.I.; Guvendik, G. N-acetyl-l-cysteine protects liver and kidney against chromium(VI)-induced oxidative stress in mice. Biol. Trace Elem. Res. 2017, 178, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Linos, A.; Petralias, A.; Christophi, C.A.; Christoforidou, E.; Kouroutou, P.; Stoltidis, M.; Veloudaki, A.; Tzala, E.; Makris, K.C.; Karagas, M.R. Oral ingestion of hexavalent chromium through drinking water and cancer mortality in an industrial area of greece—An ecological study. Environ. Health 2011, 10, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaumont, J.J.; Sedman, R.M.; Reynolds, S.D.; Sherman, C.D.; Li, L.H.; Howd, R.A.; Sandy, M.S.; Zeise, L.; Alexeeff, G.V. Cancer mortality in a chinese population exposed to hexavalent chromium in drinking water. Epidemiology 2008, 19, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Li, Y.; Luo, L.; Xie, Y.; Zeng, M.; Wang, A.; Chen, H.; Zhong, C. Role of mitochondrial electron transport chain dysfunction in Cr(VI)-induced cytotoxicity in L-02 hepatocytes. Cell. Physiol. Biochem. 2014, 33, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nino, W.R.; Zazueta, C.; Tapia, E.; Pedraza-Chaverri, J. Curcumin attenuates Cr(VI)-induced ascites and changes in the activity of aconitase and F(1)F(0) ATPase and the ATP content in rat liver mitochondria. J. Biochem. Mol. Toxicol. 2014, 28, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Piantadosi, C.A.; Suliman, H.B. Transcriptional control of mitochondrial biogenesis and its interface with inflammatory processes. Biochim. Biophys. Acta 2012, 1820, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Wenz, T. Regulation of mitochondrial biogenesis and PGC-1α under cellular stress. Mitochondrion 2013, 13, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, A.; Venero, J.L.; Iwasawa, R.; Hankir, M.K.; Rahman, S.; Boobis, A.; Hajji, N. PGC-1α controls mitochondrial biogenesis and dynamics in lead-induced neurotoxicity. Aging 2015, 7, 629–647. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, P.J.; Auwerx, J. Regulation of PGC-1 α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011, 93, 884s–890s. [Google Scholar] [CrossRef] [PubMed]
- Raefsky, S.M.; Mattson, M.P. Adaptive responses of neuronal mitochondria to bioenergetic challenges: Roles in neuroplasticity and disease resistance. Free Radic. Biol. Med. 2017, 102, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, Q.; Guo, J.; Yuan, H.; Peng, H.; Cui, L.; Yin, J.; Zhang, L.; Zhao, J.; Li, J.; et al. Non-cytotoxic concentrations of acetaminophen induced mitochondrial biogenesis and antioxidant response in HepG2 cells. Environ. Toxicol. Pharmacol. 2016, 46, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Carraway, M.S.; Suliman, H.B.; Jones, W.S.; Chen, C.W.; Babiker, A.; Piantadosi, C.A. Erythropoietin activates mitochondrial biogenesis and couples red cell mass to mitochondrial mass in the heart. Circ. Res. 2010, 106, 1722–1730. [Google Scholar] [CrossRef] [PubMed]
- Kelly, D.P.; Scarpulla, R.C. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004, 18, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Li, P.A.; Hou, X.; Hao, S. Mitochondrial biogenesis in neurodegeneration. J. Neurosci. Res. 2017, 95, 2025–2029. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, Y.; Kaguni, L.S. Matrix proteases in mitochondrial DNA function. Biochim. Biophys. Acta 2012, 1819, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Roh, Y.S.; Seki, E. Toll-like receptors in alcoholic liver disease, non-alcoholic steatohepatitis and carcinogenesis. J. Gastroenterol. Hepatol. 2013, 28, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Lu, M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med. Microbiol. Immunol. 2015, 204, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Suliman, H.B.; Sweeney, T.E.; Withers, C.M.; Piantadosi, C.A. Co-regulation of nuclear respiratory factor-1 by NFκB and CREB links LPS-induced inflammation to mitochondrial biogenesis. J. Cell Sci. 2010, 123, 2565–2575. [Google Scholar] [CrossRef] [PubMed]
- Bocco, B.M.; Louzada, R.A.; Silvestre, D.H.; Santos, M.C.; Anne-Palmer, E.; Rangel, I.F.; Abdalla, S.; Ferreira, A.C.; Ribeiro, M.O.; Gereben, B.; et al. Thyroid hormone activation by type 2 deiodinase mediates exercise-induced peroxisome proliferator-activated receptor-gamma coactivator-1α expression in skeletal muscle. J. Physiol. 2016, 594, 5255–5269. [Google Scholar] [CrossRef] [PubMed]
- LeBleu, V.S.; O’Connell, J.T.; Gonzalez Herrera, K.N.; Wikman, H.; Pantel, K.; Haigis, M.C.; de Carvalho, F.M.; Damascena, A.; Domingos Chinen, L.T.; Rocha, R.M.; et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014, 16, 992–1015. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.O.; Hitron, J.A.; Wang, X.; Chang, Q.; Pan, J.; Zhang, Z.; Liu, J.; Wang, S.; Lee, J.C.; Shi, X. Cr(VI) induces mitochondrial-mediated and caspase-dependent apoptosis through reactive oxygen species-mediated p53 activation in JB6 Cl41 cells. Toxicol. Appl. Pharmacol. 2010, 245, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K.; Moriwaki, K.; de Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar] [PubMed]
- Taherzadeh-Fard, E.; Saft, C.; Akkad, D.A.; Wieczorek, S.; Haghikia, A.; Chan, A.; Epplen, J.T.; Arning, L. PGC-1α downstream transcription factors NRF-1 and TFAM are genetic modifiers of huntington disease. Mol. Neurodegener. 2011, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Cruzat, V.F.; Newsholme, P.; Cheng, J.; Chen, Y.; Lu, Y. Regulation of SIRT1 in aging: Roles in mitochondrial function and biogenesis. Mech. Ageing Dev. 2016, 155, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, S.; Li, J.; Zheng, L.; Feng, M.; Wang, X.; Han, K.; Pi, H.; Li, M.; Huang, X.; et al. SIRT1 facilitates hepatocellular carcinoma metastasis by promoting PGC-1α-mediated mitochondrial biogenesis. Oncotarget 2016, 7, 29255–29274. [Google Scholar] [CrossRef] [PubMed]
- Kitagishi, Y.; Matsuda, S. Redox regulation of tumor suppressor pten in cancer and aging (review). Int. J. Mol. Med. 2013, 31, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Tobita, T.; Guzman-Lepe, J.; Takeishi, K.; Nakao, T.; Wang, Y.; Meng, F.; Deng, C.X.; Collin de l’Hortet, A.; Soto-Gutierrez, A. SIRT1 disruption in human fetal hepatocytes leads to increased accumulation of glucose and lipids. PLoS ONE 2016, 11, e0149344. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Rhee, J.; Donovan, J.; Walkey, C.J.; Yoon, J.C.; Oriente, F.; Kitamura, Y.; Altomonte, J.; Dong, H.; Accili, D.; et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 2003, 423, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Olmos, Y.; Valle, I.; Borniquel, S.; Tierrez, A.; Soria, E.; Lamas, S.; Monsalve, M. Mutual dependence of FOXO3a and PGC-1α in the induction of oxidative stress genes. J. Biol. Chem. 2009, 284, 14476–14484. [Google Scholar] [CrossRef] [PubMed]
- Daitoku, H.; Sakamaki, J.; Fukamizu, A. Regulation of FOXO transcription factors by acetylation and protein-protein interactions. Biochim. Biophys. Acta 2011, 1813, 1954–1960. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Liu, Y.; Gao, R.; Li, H.; Dunn, T.; Wu, P.; Smith, R.G.; Sarkar, P.S.; Fang, X. Ethanol suppresses PGC-1α expression by interfering with the camp-creb pathway in neuronal cells. PLoS ONE 2014, 9, e104247. [Google Scholar] [CrossRef] [PubMed]
- Valero, T. Mitochondrial biogenesis: Pharmacological approaches. Curr. Pharm. Des. 2014, 20, 5507–5509. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Li, L.C.; Chen, J.B.; Chang, H.W. Indoxyl sulfate-induced oxidative stress, mitochondrial dysfunction, and impaired biogenesis are partly protected by vitamin C and N-acetylcysteine. Sci. World J. 2015, 2015, 620826. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, R.; Satish Kumar, C.S.; Rani, M.U.; Srikanth, M.K.; Boobalan, G.; Reddy, A.G. An evaluation of the protective role of α-tocopherol on free radical induced hepatotoxicity and nephrotoxicity due to chromium in rats. Indian J. Pharmacol. 2013, 45, 490–495. [Google Scholar] [PubMed]
- Arreola-Mendoza, L.; Reyes, J.L.; Melendez, E.; Martin, D.; Namorado, M.C.; Sanchez, E.; Del Razo, L.M. α-Tocopherol protects against the renal damage caused by potassium dichromate. Toxicology 2006, 218, 237–246. [Google Scholar] [PubMed]
- Kumar, D.; Gangwar, S.P. Role of antioxidants in detoxification of Cr(VI) toxicity in laboratory rats. J. Environ. Sci. Eng. 2012, 54, 441–446. [Google Scholar] [PubMed]
- Olguin-Martinez, M.; Hernandez-Espinosa, D.R.; Hernandez-Munoz, R. α-Tocopherol administration blocks adaptive changes in cell NADH/NAD+ redox state and mitochondrial function leading to inhibition of gastric mucosa cell proliferation in rats. Free Radic. Biol. Med. 2013, 65, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Truksa, J.; Dong, L.F.; Rohlena, J.; Stursa, J.; Vondrusova, M.; Goodwin, J.; Nguyen, M.; Kluckova, K.; Rychtarcikova, Z.; Lettlova, S.; et al. Mitochondrially targeted vitamin e succinate modulates expression of mitochondrial DNA transcripts and mitochondrial biogenesis. Antioxid. Redox Signal. 2015, 22, 883–900. [Google Scholar] [CrossRef] [PubMed]
- Luczak, M.W.; Zhitkovich, A. Role of direct reactivity with metals in chemoprotection by N-acetylcysteine against chromium(VI), cadmium(II), and cobalt(II). Free Radic. Biol. Med. 2013, 65, 262–269. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| PGC-1a | TCTGTGTCACTGTGGATTGGAG | AGTTCAGGAAGATCTGGGCAA |
| Tfam | TGCGGTTTCCCTTCATCTCC | ACTAGCGAGGCACTATGGGA |
| NRF-1 | CCTGGTCCAGAACTTTACACAGA | CACTCCGTGTTCCTCCATGA |
| NRF-2 | AACCAGTGGATCTGCCAACT | AAGTGACTGAAACGTAGCCG |
| SIRT1 | GCGGTTCCTACTGCGCGA | TCACTAGAGCTTGCATGTGAGG |
| MAPK1 | GCCGAAGCACCATTCAAGTT | GATGTCTGAGCACGTCCAGT |
| mtDNA | CAAACCTACGCCAAAATCCA | GAAATGAATGAGCCTACAGA |
| AKT1 | CAGGATGTGGACCAACGTGA | AAGGTACGTTCGATGACAGT |
| CREB1 | TTCAAGCCCAGCCACAGATT | AGTTGAAATCTGAACTGTTTGGAC |
| PTEN | CTCCCAGACATGACAGCCA | GCTTTGAATCCAAAAACCTTACTCA |
| Caspase3 | TGGTTTTCGGTGGGTGTG | CCACTGAGTTTTCAGTGTTCTC |
| Caspaase8 | AGTCTGTACCTTTCTGGCGG | TCTCCCAGGATGACCCTCTT |
| GAPDH | TGACAACAGCCTCAAGAT | GAGTCCTTCCACGATACC |
| Assay Name | Assay ID | Catalog Number |
|---|---|---|
| 18S | Hs99999901 | 4331182 |
| ATP5J | Hs01081394_g1 | 4331182 |
| ATP5O | Hs00919163 | 4331182 |
| NDUFA1 | Hs00915157_g1 | 4331182 |
| NDUFB1 | Hs00929425 | 4331182 |
| TLR2 | Hs02621280_s1 | 4331182 |
| TLR4 | Hs00152939_m1 | 4331182 |
| TNF | Hs01113624_g1 | 4331182 |
| IL-1β | Hs01555410_m1 | 4331182 |
| SOD1 | Hs00533490_m1 | 4331182 |
| SOD2 | Hs00167309_m1 | 4331182 |
| GSTO1 | Hs02383465_s1 | 4331182 |
| FOXO1 | Hs00231106_m1 | 4331182 |
| KEAP1 | hs01003430 | 4331182 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, X.; De Cássia da Silveira e Sá, R.; Zhong, C. Mitochondrial Biogenesis in Response to Chromium (VI) Toxicity in Human Liver Cells. Int. J. Mol. Sci. 2017, 18, 1877. https://doi.org/10.3390/ijms18091877
Zhong X, De Cássia da Silveira e Sá R, Zhong C. Mitochondrial Biogenesis in Response to Chromium (VI) Toxicity in Human Liver Cells. International Journal of Molecular Sciences. 2017; 18(9):1877. https://doi.org/10.3390/ijms18091877
Chicago/Turabian StyleZhong, Xiali, Rita De Cássia da Silveira e Sá, and Caigao Zhong. 2017. "Mitochondrial Biogenesis in Response to Chromium (VI) Toxicity in Human Liver Cells" International Journal of Molecular Sciences 18, no. 9: 1877. https://doi.org/10.3390/ijms18091877





